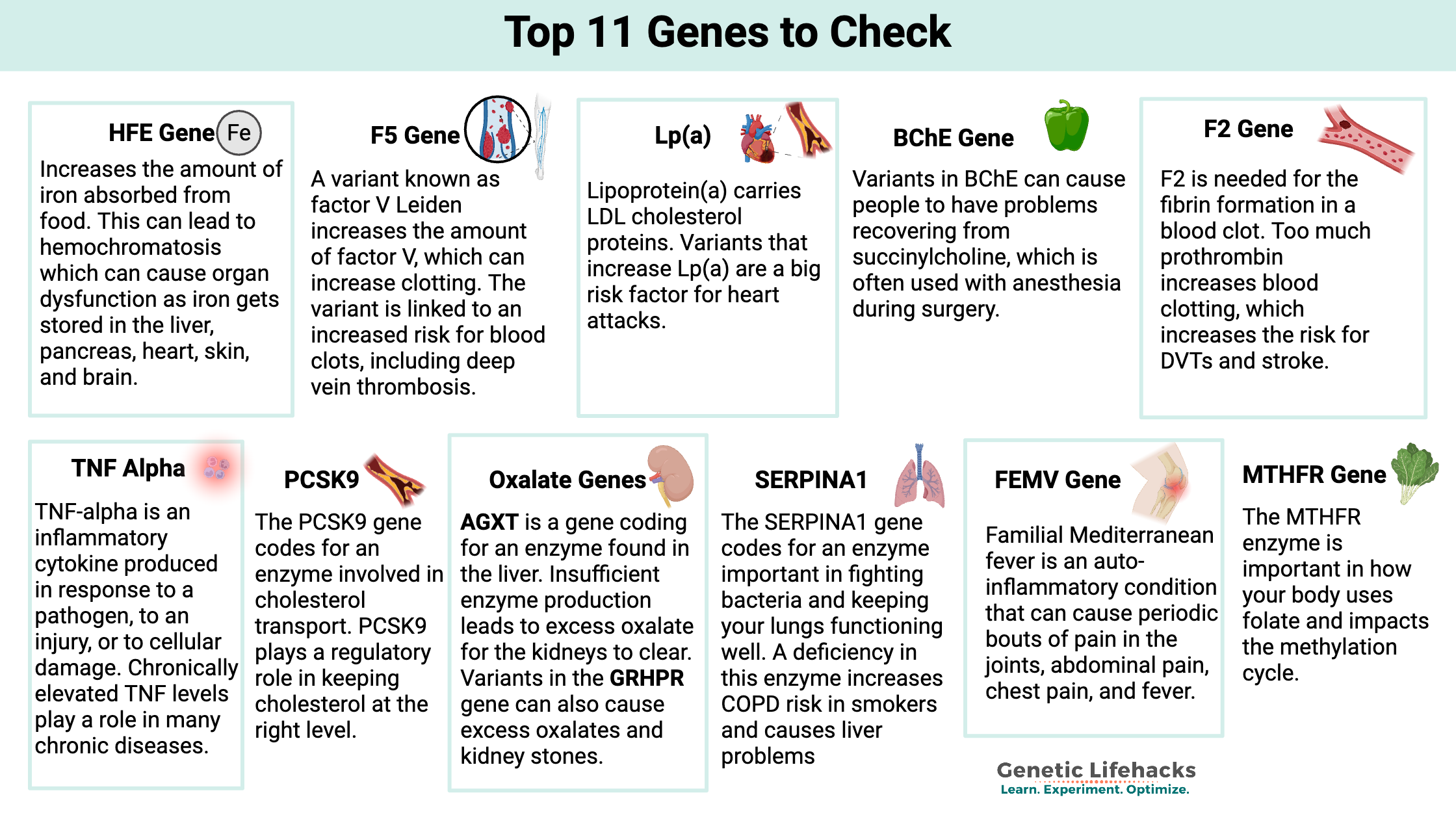
Getting started with learning about your genes can be overwhelming! Let me help you cut through the information overload. This guide offers a research-backed shortlist of genetic variants that can directly impact health and wellness.
Instead of overwhelming you with data, I’m focusing on 11 of the most clinically relevant genes that are found in 23andMe and AncestryDNA raw data. These are genes with robust research showing significant, actionable health risks or traits.
For each gene, you’ll learn:
-
Why it matters: A brief summary of its clinical importance
-
Which variants (SNPs) to look at: Members will see their genotypes right in the report, with highlighting to indicate a risk
-
Where to get more information: If you have a highlighted variant, you can find more details, including what to talk with your doctor about and possible natural solutions.
Top 11 List: Genes and SNPs To Check in Your Raw Data
If you have a risk variant highlighted below, be sure to click through to read the full article for in-depth background science and the “Lifehacks” that may help mitigate any negative effects. Knowledge is power here, and understanding your genes can help you prevent and avoid health issues.
#1) HFE Gene (hemochromatosis risk)
Access this content:
An active subscription is required to access this content.
References:
Chasman, Daniel I., et al. “Polymorphism in the Apolipoprotein(a) Gene, Plasma Lipoprotein(a), Cardiovascular Disease, and Low-Dose Aspirin Therapy.” Atherosclerosis, vol. 203, no. 2, Apr. 2009, pp. 371–76. PubMed, https://doi.org/10.1016/j.atherosclerosis.2008.07.019.
Chen, Suet N., et al. “A Common PCSK9 Haplotype, Encompassing the E670G Coding Single Nucleotide Polymorphism, Is a Novel Genetic Marker for Plasma Low-Density Lipoprotein Cholesterol Levels and Severity of Coronary Atherosclerosis.” Journal of the American College of Cardiology, vol. 45, no. 10, May 2005, pp. 1611–19. PubMed, https://doi.org/10.1016/j.jacc.2005.01.051.
Erhart, Gertraud, et al. “Genetic Factors Explain a Major Fraction of the 50% Lower Lipoprotein(a) Concentrations in Finns.” Arteriosclerosis, Thrombosis, and Vascular Biology, vol. 38, no. 5, May 2018, pp. 1230–41. PubMed Central, https://doi.org/10.1161/ATVBAHA.118.310865.
Fargue, Sonia, et al. “Four of the Most Common Mutations in Primary Hyperoxaluria Type 1 Unmask the Cryptic Mitochondrial Targeting Sequence of Alanine:Glyoxylate Aminotransferase Encoded by the Polymorphic Minor Allele.” The Journal of Biological Chemistry, vol. 288, no. 4, Jan. 2013, pp. 2475–84. PubMed, https://doi.org/10.1074/jbc.M112.432617.
Gomes, Henrique J. P., et al. “Investigation of Association between Susceptibility to Leprosy and SNPs inside and near the BCHE Gene of Butyrylcholinesterase.” Journal of Tropical Medicine, vol. 2012, 2012, p. 184819. PubMed, https://doi.org/10.1155/2012/184819.
Lando, Giuliana, et al. “Frequency of Butyrylcholinesterase Gene Mutations in Individuals with Abnormal Inhibition Numbers: An Italian-Population Study.” Pharmacogenetics, vol. 13, no. 5, May 2003, pp. 265–70. PubMed, https://doi.org/10.1097/00008571-200305000-00005.
Laschkolnig, Anja, et al. “Lipoprotein (a) Concentrations, Apolipoprotein (a) Phenotypes, and Peripheral Arterial Disease in Three Independent Cohorts.” Cardiovascular Research, vol. 103, no. 1, July 2014, pp. 28–36. PubMed, https://doi.org/10.1093/cvr/cvu107.
NM_000055.2(BCHE):C.293A>G (p.Asp98Gly) AND Deficiency of Butyrylcholine Esterase – ClinVar – NCBI. https://www.ncbi.nlm.nih.gov/clinvar/RCV000277104.1/. Accessed 19 May 2022.
NM_012203.1(GRHPR):C.404+3_404+6delAAGT AND Primary Hyperoxaluria, Type II – ClinVar – NCBI. https://www.ncbi.nlm.nih.gov/clinvar/RCV000186457.2/. Accessed 19 May 2022.
Pezzini, Alessandro, et al. “Do Common Prothrombotic Mutations Influence the Risk of Cerebral Ischaemia in Patients with Patent Foramen Ovale? Systematic Review and Meta-Analysis.” Thrombosis and Haemostasis, vol. 101, no. 5, May 2009, pp. 813–17.
Qiu, Chengfeng, et al. “What Is the Impact of PCSK9 Rs505151 and Rs11591147 Polymorphisms on Serum Lipids Level and Cardiovascular Risk: A Meta-Analysis.” Lipids in Health and Disease, vol. 16, no. 1, June 2017, p. 111. PubMed, https://doi.org/10.1186/s12944-017-0506-6.
Slimani, Afef, et al. “Effect of E670G Polymorphism in PCSK9 Gene on the Risk and Severity of Coronary Heart Disease and Ischemic Stroke in a Tunisian Cohort.” Journal of Molecular Neuroscience: MN, vol. 53, no. 2, June 2014, pp. 150–57. PubMed, https://doi.org/10.1007/s12031-014-0238-2.
Thun, Gian Andri, et al. “Causal and Synthetic Associations of Variants in the SERPINA Gene Cluster with Alpha1-Antitrypsin Serum Levels.” PLoS Genetics, vol. 9, no. 8, Aug. 2013, p. e1003585. PubMed Central, https://doi.org/10.1371/journal.pgen.1003585.
Zhu, Guang-dan, et al. “Genetic Testing for BCHE Variants Identifies Patients at Risk of Prolonged Neuromuscular Blockade in Response to Succinylcholine.” Pharmacogenomics and Personalized Medicine, vol. 13, Sept. 2020, pp. 405–14. PubMed Central, https://doi.org/10.2147/PGPM.S263741.
Debbie Moon is a biologist, engineer, author, and the founder of Genetic Lifehacks where she has helped thousands of members understand how to apply genetics to their diet, lifestyle, and health decisions. With more than 10 years of experience translating complex genetic research into practical health strategies, Debbie holds a BS in engineering from Colorado School of Mines and an MSc in biological sciences from Clemson University. She combines an engineering mindset with a biological systems approach to explain how genetic differences impact your optimal health.