Do you have both AncestryDNA and 23andMe raw data files? If so, you can combine the raw data files into one file to use with your Genetic Lifehacks membership.
4 Ways to Combine Data Files:
There are several ways to combine 23andMe and AncestryDNA data files — the easy way and the hard way. The hard way is a nifty solution that works on Macs and Linux.
1) The Easy Way to Combine Data Files (Windows or Mac):
- Open your unzipped 23andMe data file in Notepad (Windows) or TextEdit (Mac). Save the file with a new name – e.g., combined_data_file.txt. Make sure that you save it as a .txt file.
- Next, open your AncestryDNA data file in the text editor. Copy all of the AncestryDNA data file. [Select all by pressing Ctrl-A (Windows) or Cmd-A (Mac). Then copy by pressing Ctrl-C (Windows) or Cmd-C (Mac).]
- Go back to the file you saved with the new name (e.g., combined_data_file.txt) and paste the AncestryDNA data at the end of your 23andMe data.
- Save the file.
- Go to the Member Dashboard and click the “Clear Data” button. Then click the “Choose File” button and connect to your combined data file.
That’s it! Quick and simple.
2) The harder way to combine the data file (removes the duplicates, smaller file size)
This method of combining the 23andMe and AncestryDNA raw data files involves using a Mac or Linux terminal window.
These directions assume you have some minor skills in navigating a terminal window.
Download first: First, be sure you have downloaded both the 23andMe and AncestryDNA raw data files. Know where the .txt files are on your hard drive, and make sure they are both in the same folder.
Open a terminal window and navigate to the folder where your raw data files are located.
1 ) Strip out the header information on both text files.
Before we combine the two files, let’s clean up the extra information at the top of each file. Simply open the files in TextEdit (or your default .txt file editor) and delete the first lines that explain how the file was created.
It will look like this:
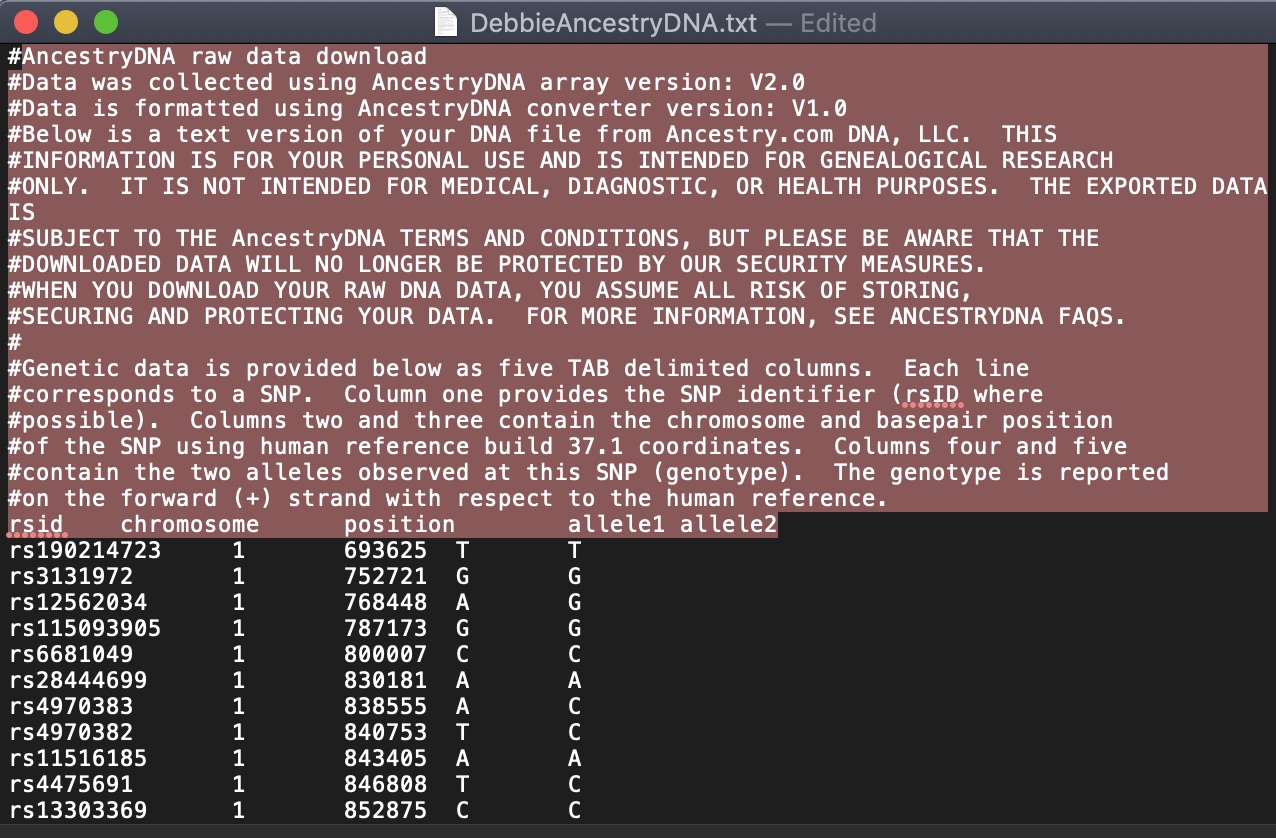
Save the files and close the text editor.
2) Combine alleles 1 and 2 on the AncestryDNA file:
The data for the AncestryDNA file is given in five columns – rs id, chromosome #, position, and then allele 1 and allele 2.
The data from 23andMe is given in four columns – rs id, chromosome #, position, and then combined genotype (allele1 + allele2).
Thus, we need to make the AncestryDNA file (5 columns) match with the 23andMe data (4 columns). To do this, in the terminal window, we will run an AWK command combining the 4th and 5th columns in the AncestryDNA data file. This will output to a new file name.
Be sure to change the filenames to whatever you named your 23andMe and AncestryDNA files.
awk 'BEGIN {FS="\t"};{print $1"\t"$2"\t"$3"\t"$4 and $5}' AncestryDNA.txt > AncestryCombined.txt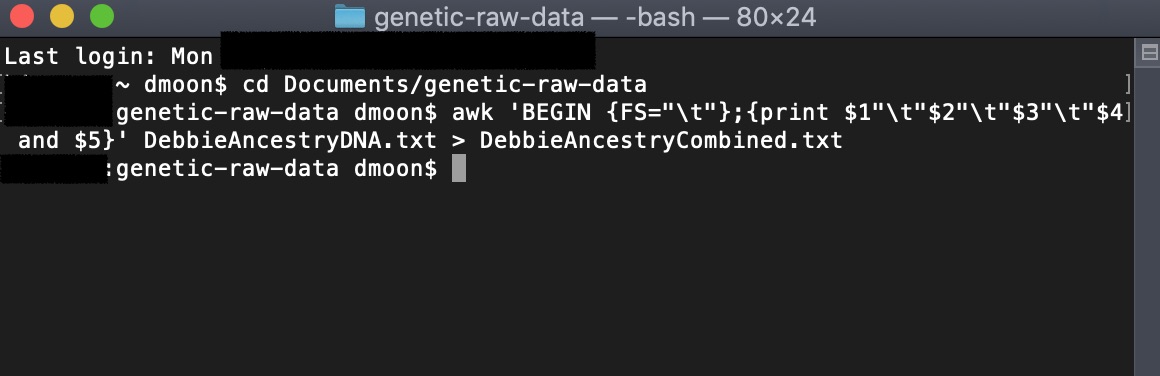
Now your AncestryDNA file will be in the same format as the 23andMe raw data file (4 columns, genotype combined).
3) Combine the 23andMe and the edited AncestryDNA files, and remove duplicates:
We will use a couple more commands to combine the two files and remove the duplicate rs ids.
Again, be sure to change the file names to whatever your file names are.
cat 23andMe.txt AncestryCombined.txt | awk '!seen[$0]++' > ComboRawGenome.txt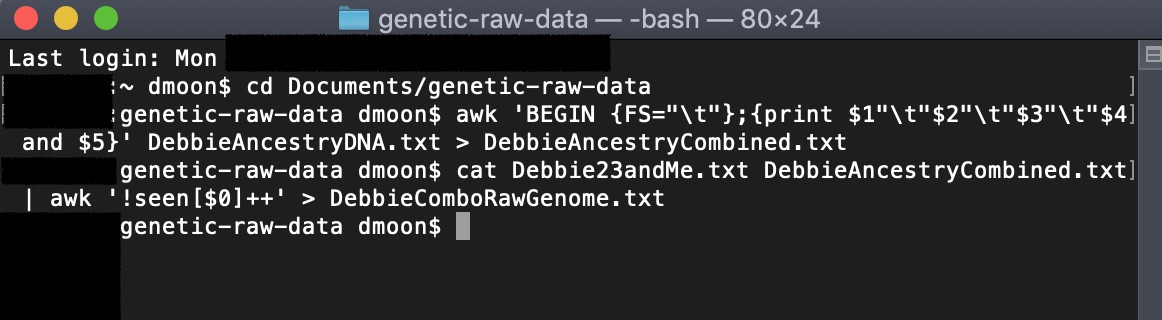
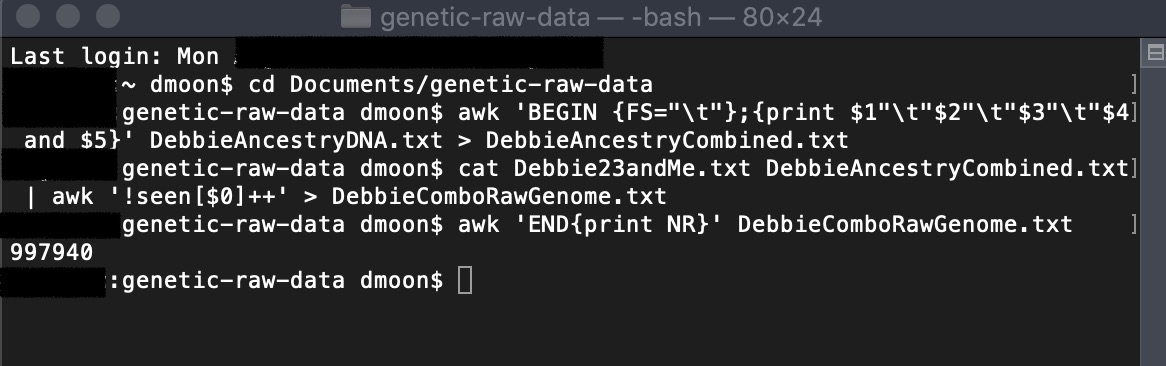
If you open up your newly created file (ComboRawGenome.txt), you will see all of your genetic data combined. The original files for 23andMe v4, v5, and AncestryDNA are between 600,000 to 700,000 rows (depending on when you did the tests). You can check to see how many rows are in the combined files using the following awk command:
awk 'END{print NR}' ComboRawGenome.txtThe number it outputs is the number of rows. In my example, I was using 23andMe v4 and AncestryDNA v2 data with the mitochondrial and Y chromosome data stripped out to give me a combined total of 997, 940 rows (which can be imported into Excel).
That’s it! Now you have a file with the combined 23andMe and Ancestry data with duplicates removed. Note, in the example above, I put the 23andMe data file first. For the final file, the first instance of the rs id (in this case, 23andMe data) will be included, and the second instance of the rs id (AncestryDNA) will be removed.
Now go to the Member Dashboard and connect to your new combined data file.
Option #3) A little more complicated but with even cleaner data
A Genetic Lifehacks member came up with an even better solution using AWK to clean up the data first and strip out any 00 or — that are in the data files from AncestryDNA and 23andMe.
Here are the step-by-step directions on his blog: https://jamesalvarez.co.uk/blog/genome/
Option 4) Python script
If you prefer Python, here’s another option for combining the data files and cleaning up the data: https://github.com/michael-patzer/combine-dna