To use your Dante Labs whole-genome file on Genetic Lifehacks, you will need to convert the data into a format that looks like 23 and Me or AncestryDNA raw data.
This article covers one way to convert the whole genome BAM to a .txt file in the 23andMe format using a free software tool. I recommend doing this on a desktop or fast laptop computer (not a clunky, old laptop or Chromebook…).
Quick note: A member alerted me to DNAgenics, which will do the conversion for you for a fee.
Download your whole genome file from Dante Labs:
1.) Log in to Dante Labs
Go to genome.dantelabs.com and log in.
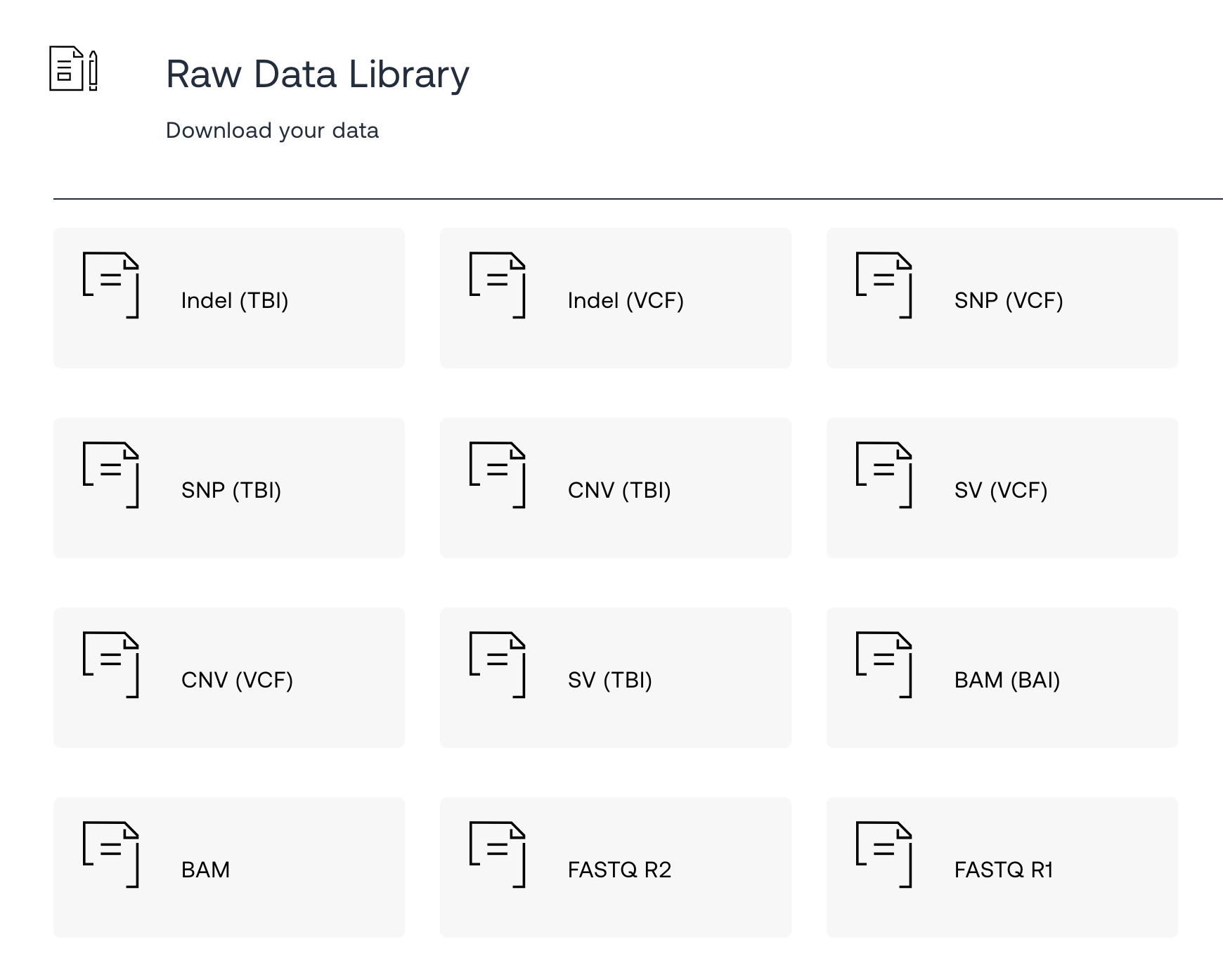
2.) Under the Reports dropdown, choose Raw Data Library
3.) You’ll have a lot of options for different file formats. Click on BAM to download the BAM file. It is about a 50GB file, so downloading it may take a while. (When I went to download my file just now, the file didn’t exist. I had to contact Dante Labs support about the error. Sigh. Hopefully, it goes smoothly for you. ***Update from a member: Apparently, you need to download the files within a few months of getting your genome sequenced. Otherwise, they go into storage, and you may have to pay a subscription fee to access the BAM file. )
Converting the BAM file using WGS Extract:
To convert the whole genome BAM file, I used WGS Extract, which is free, open-source software:
https://github.com/WGSExtract/WGSExtract.github.io
A detailed instruction manual is available on the GitHub page (100+ pages).
Yep, this is one of those times that you will need to read the manual. The software is a bit rough around the edges regarding user interface and installation. The plus side is that the application is free, works well (I’m on a Mac), and did exactly what I needed.
Follow the installation instructions for your operating system.
Note: For the reference library, use hs37d5. It was option 13 in WGSE v3.
Once WGSE is installed and running:
1.) Select your BAM file.
2.) When you load in your Dante Labs BAM file, it will pop up an alert that it needs to be indexed.
Click on the Index button next to where it says Statistics and Attributes:
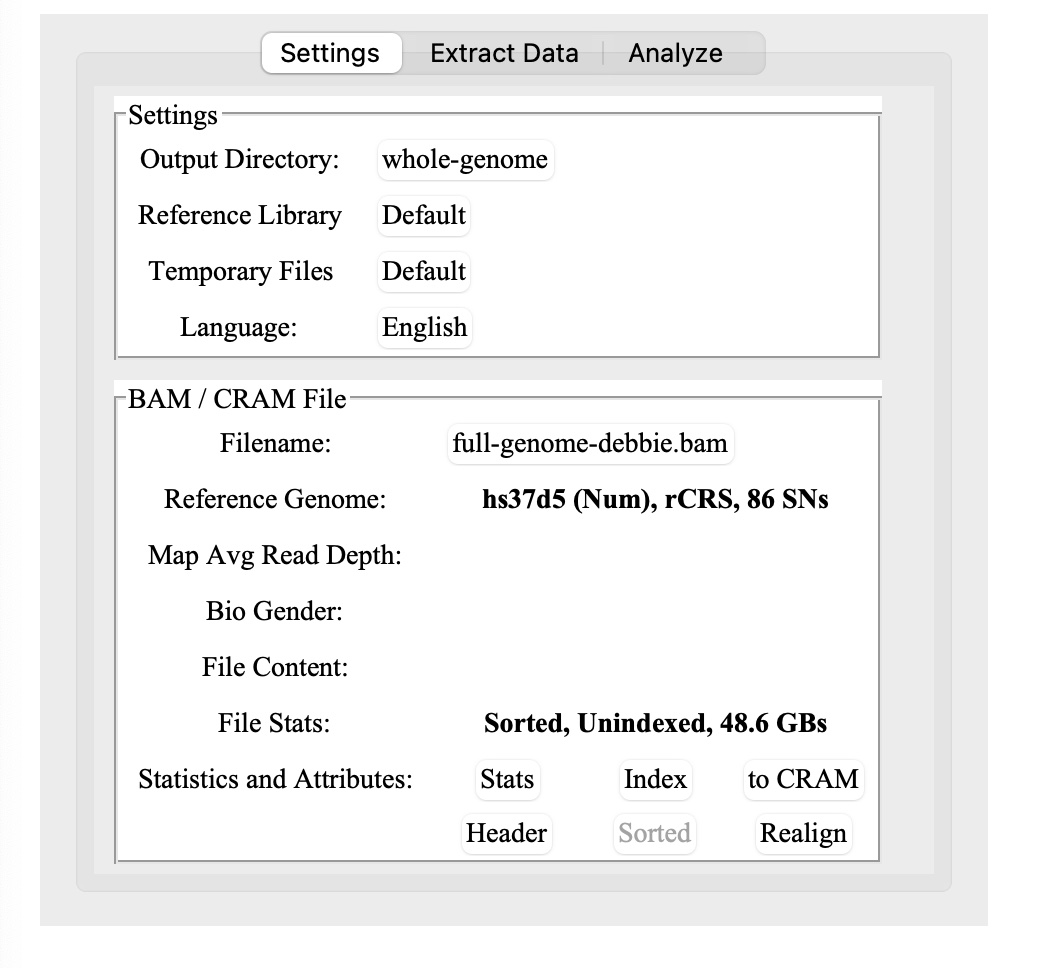
3.) Now, wait 30 minutes or so to generate the BAM index file. (It didn’t take quite that long on my Mac.) You can do other things on your computer while waiting, but don’t close the application or the terminal window.
For example, you can read through the manual again while you wait… :-)
4.) Key thing: once you’ve indexed the BAM file, you will need to click on the Stats button before you can do anything else. Yes, it clearly says this in the manual, but I missed it and was confused for a bit.
5.)Next, click on “Extract Data” and then on the “Microarray RAW” button.
To use the Genetic Lifehacks membership features, either choose 23 and Me v4, v5, and Ancestry v2 — or select Combined file. There are many other options to come back later and play with here.
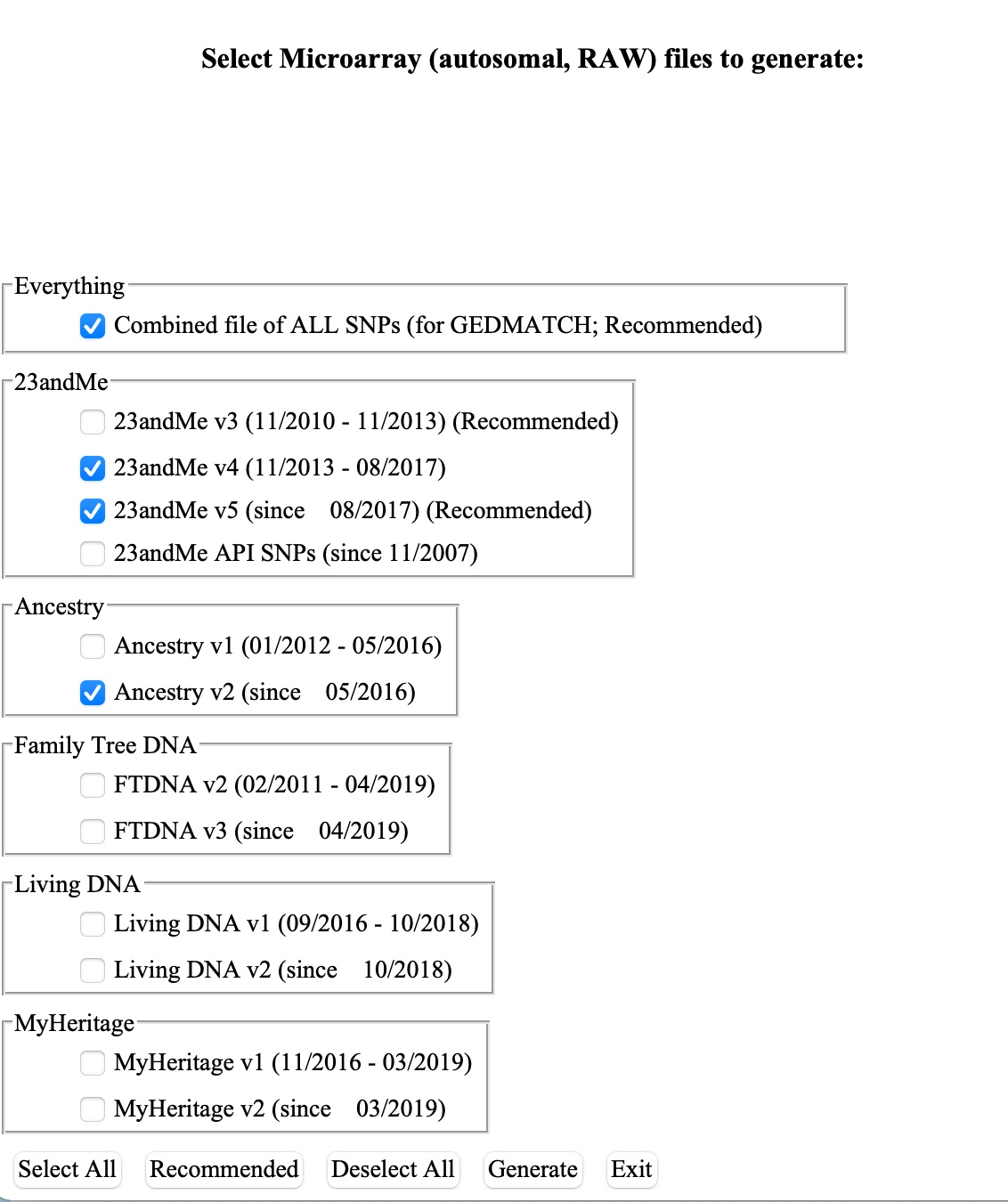
6. Click the Generate button.
It will give you an expected wait – mine said 50 minutes, but it took about half that.
Your data files should now be in the folder that you set up for WGSE to use.
7. That’s it! You should be able to connect on the Member’s Start page to the new raw data file. If you are already using a different raw data file, you must first hit the “Clear Data” button before connecting to the new data file.

