Key takeaways:
~ Many healthy foods contain oxalates, but dietary intake must be balanced with the metabolism and excretion of oxalates.
~ Genetic variants can disrupt the balance of oxalates, leading to the formation of kidney stones or oxalate crystals.
~ Genes also impact the biosynthesis of oxalates in the body.
Members will see their genotype report below and the solutions in the Lifehacks section. Consider joining today.
Oxalates, Kidney Stones, Joint Pain
Listen to this section of the article:
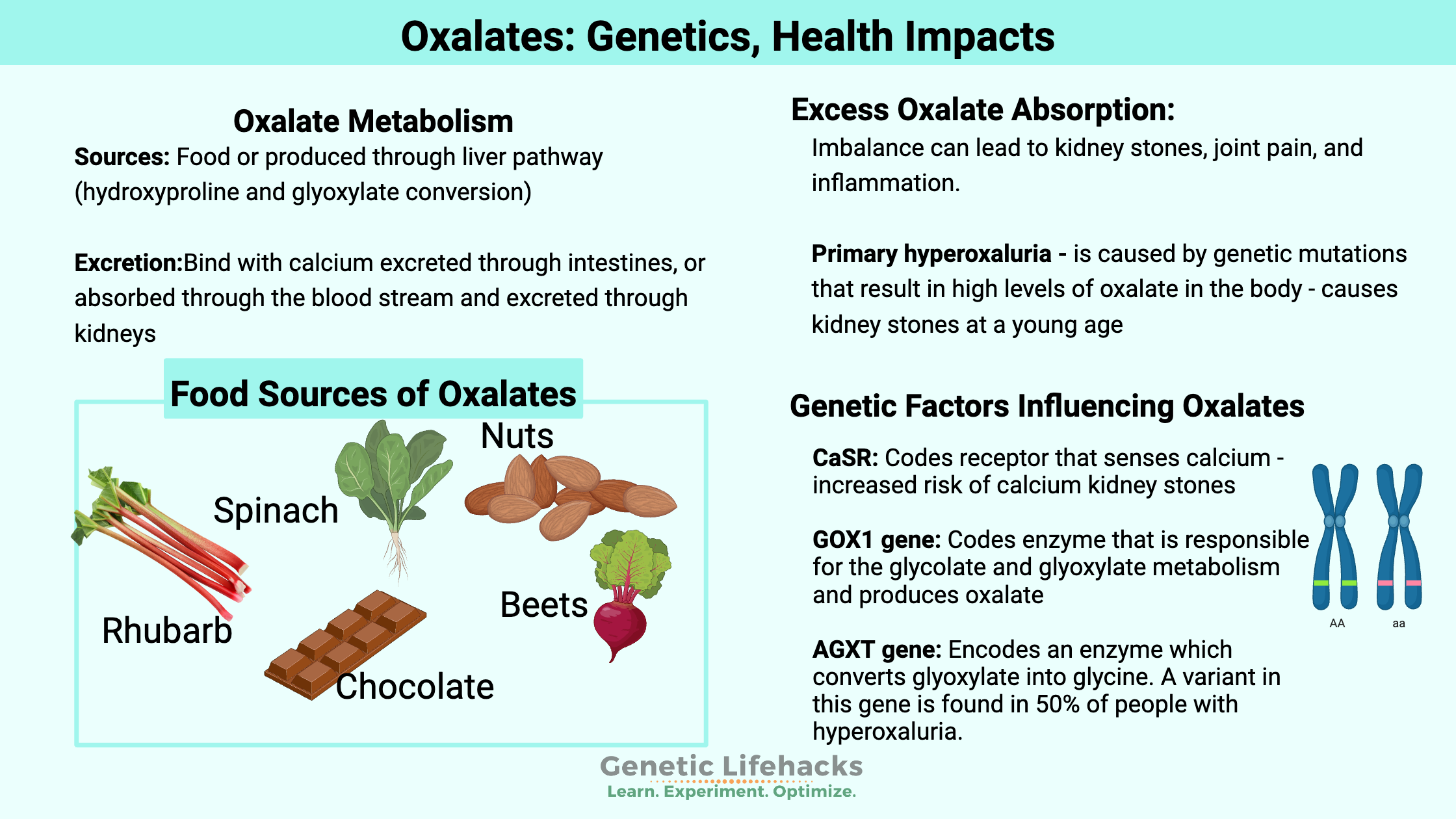
Oxalates are organic compounds found in many of the foods we eat. We can also produce oxalates in the body through certain pathways.
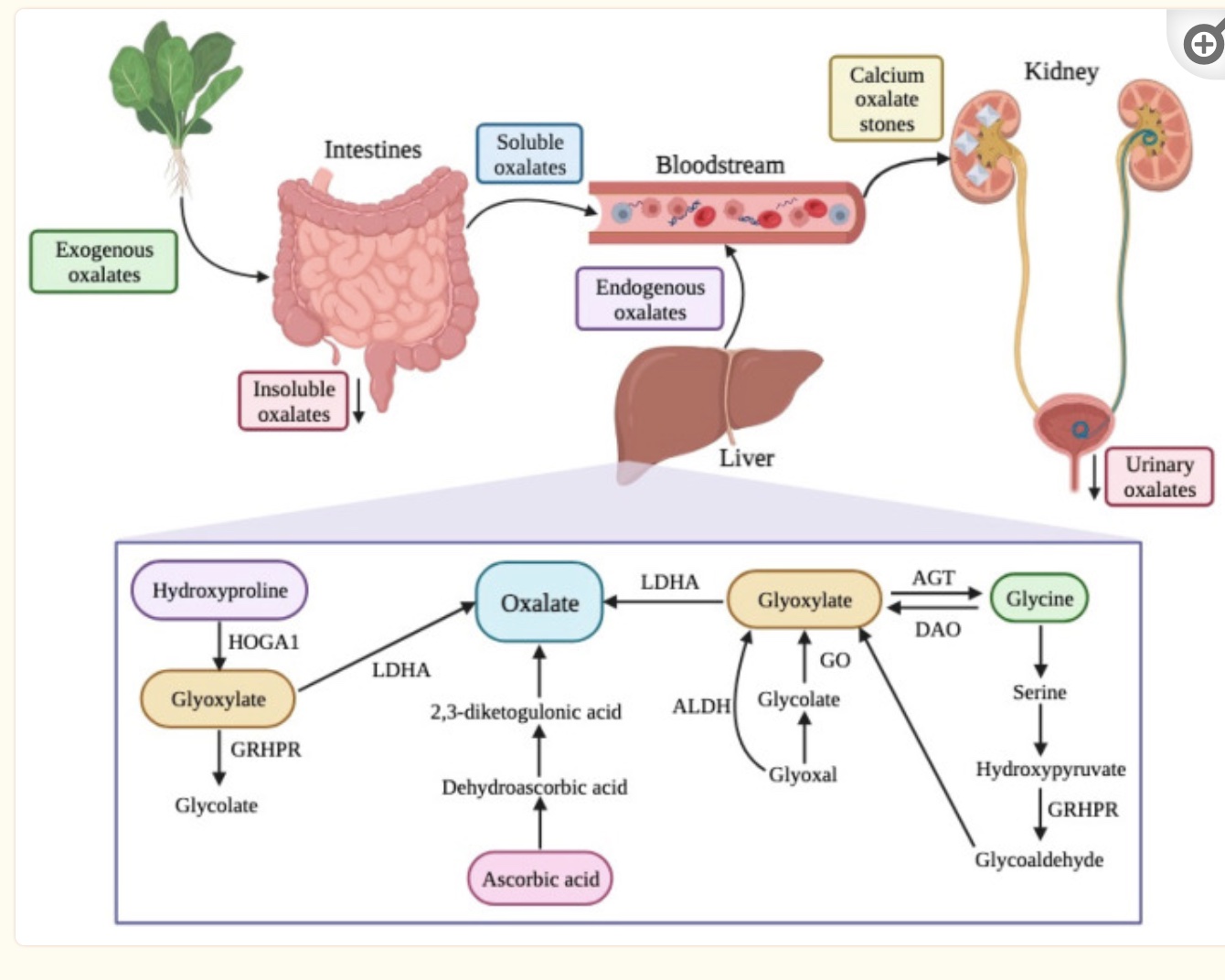
Oxalates can bind with calcium and be excreted through the intestines, or they can enter the bloodstream and eventually be excreted through the kidneys.
The levels of oxalates in the body need to balance with the metabolism and excretion matching the production and intake. When this is out of balance, problems can arise with oxalates causing kidney stones or other joint problems in some people.
Where do oxalates come from?
We get oxalates from foods such as spinach, rhubarb, chocolate, beets, and nuts, and we can make oxalates in our bodies.
Oxalates are found in fruits and vegetables – some fruits and vegetables contain high levels of oxalates, while others contain only a tiny amount. When we eat plants high in oxalates, the gut microbiome takes care of eliminating some of the oxalates, and then our intestines absorb some of the oxalates we have eaten. Oxalates are absorbed as free oxalate. If calcium is available in the intestines (e.g. from consuming dairy products), it easily binds to oxalates and prevents absorption.[ref]
Our bodies also produce different forms of oxalate as they metabolize different substances. The biosynthesis of oxalates in the liver accounts for 50-80% of the body’s oxalate pool. Dietary intake accounts for the remainder.[ref]
Precursors of oxalates produced in the body include amino acids and carbohydrate sources. Hydroxyproline, from collagen breakdown, and glycine are amino acid sources that can be metabolized to form oxalates. Glycolate, glyoxylate, and glyoxal are derived from carbohydrates and can form oxalates in the body through their catabolic pathway.[ref]
How are oxalates eliminated?
When you eat foods that contain oxalates, your gut microbiome plays a big role in how much of the oxalates you will absorb. The bacteria Oxalobacter formigenes uses oxalates as a primary energy source, so the presence of this bacteria can protect against oxalate absorption. One study found that people who had Oxalobacter formigenes in their gut microbiome had lower urinary oxalate levels. The same study found that the abscence of Oxalobacter formigenes correlated with higher plasma oxalate levels.[ref]
The kidneys play a key role in maintaining the balance of oxalates in the body. Free oxalates are excreted by the kidneys, or they can combine with calcium ions in the kidneys to form calcium oxalate crystals. If the calcium oxalate crystals infiltrate the vessel walls in the kidneys, this can lead to the formation of kidney stones or even kidney disease.[ref]
In the kidneys and liver, the amino acid hydroxyproline is metabolized in the mitochondria to form pyruvate (used for ATP production) and glyoxylate. Hydroxyproline can be catabolized from collagen, such as that found in your connective tissue. The normal daily turnover of collagen in the body is estimated to be 2-3 g/day. [ref] The AGXT gene creates the enzyme responsible for breaking down glyoxylate into glycine so that it doesn’t get converted into oxalates.
High oxalate levels:
There is a delicate balance of oxalate levels maintained by the absorption of oxalate from food, the production of oxalate in the liver, and the filtration and secretion or reabsorption of oxalate in the kidneys.
If any of these pieces go awry — gut microbiome issues, too much absorption in the gut, kidney disease, genetic variants that shift you towards making more oxalates — you can end up with high oxalate levels.
Kidney stone:
High levels of oxalate in the kidneys can lead to the formation of calcium oxalate kidney stones.[ref] Approximately 80% of kidney stones are composed of oxalate bound to calcium. According to a 2005 study, “5% of American women and 12% of men will develop a kidney stone at some time in their life, and prevalence has been rising in both sexes.”[ref]
Urinary nanocrystals:
Urinary crystals can form in urine that contains calcium and oxalates. These may be the precursors to kidney stones in people with certain changes in their kidney morphology. A study in healthy adults showed that a low-oxalate diet decreased urinary oxalate nanocrystals and a high-oxalate diet significantly increased urinary oxalate nanocrystals. [ref]
Oxalate crystals:
In addition to forming kidney stones, oxalate crystals can sometimes be deposited in the joints, skin, and retina.[ref][ref] This can cause joint pain and inflammation.
Research on oxalate-induced arthritis, or joint inflammation, says that it is an uncommon cause of arthritis. It is caused by the deposition of calcium oxalate crystals in the synovial fluid that surrounds a joint. Often this type of arthritis is seen in people with genetic mutations that cause high oxalate levels.[ref][ref]
Oxalates in foods:
Foods high in oxalates include spinach, rhubarb, wheat bran, nuts, and chocolate. (See the Lifehacks section for a more complete list.)
In addition, hydroxyproline is an amino acid found in many protein-rich foods that can be metabolized into oxalates. Gelatin and collagen supplements are particularly high in hydroxyproline.[ref]
Genetics and Oxalates:
Genetic variants in genes related to oxalate formation, transport, and excretion are associated with altered oxalate levels in the body.
Many of the studies on oxalates focus on kidney stone formation, and these studies provide a clear picture of the interaction between oxalate homeostasis, kidney function, and calcium levels.
Primary hyperoxaluria (PH) is caused by genetic mutations that result in high levels of oxalate in the body. People with primary hyperoxaluria are likely to develop kidney stones at a young age.
There are three types of primary hyperoxaluria. PH1 (primary hyperoxaluria type 1) is caused by a mutation in the AGXT gene, which causes a deficiency in the body’s ability to metabolize glyoxylate into other compounds. PH2 is caused by mutations in the GRHPR gene, which is also involved in glyoxylate metabolism. PH3 is caused by mutations in the HOGA1 gene, which is involved in the breakdown of hydroxyproline in the mitochondria for use as pyruvate or glyoxalate.[ref]
Oxalates Genotype Report
Lifehacks:
Low Oxalate Diet:
If you carry one of the pathogenic hyperoxaluria genetic variants, talk with your doctor and consider whether it would be appropriate to adopt a diet lower in oxalates. At the least, be aware of high oxalate foods and how much you consume.
High oxalate foods include:[ref][ref]
- Spinach
- Chard
- Rhubarb
- Sorrel
- Amaranth
- Starfruit
- Okra
- Soybeans (raw, boiled)
- Sweet potato
- Swiss Chard
- Taro leaves
- Chocolate
- Nuts
- Beets
- Rice and Wheat Bran
- Licorice root
Here is another list: Foods that contain oxalates
Stacking calcium with high oxalate foods:
Related Articles and Topics:
Familial Mediterranean Fever
Familial Mediterranean fever (FMF) is a genetic condition of inflammatory episodes that cause painful joints, pain in the abdomen, or pain in the chest and is most often accompanied by a fever. FMF is often misdiagnosed as various pain-related conditions, such as fibromyalgia, myofascial pain syndrome, or gouty arthritis.
Hacking Irritable Bowel Syndrome Based on Your Genes
There are multiple causes of irritable bowel syndrome (IBS), and genetics can definitely play a role in IBS symptoms. Pinpointing your genetic cause may help you to figure out the right solution for you.
Gut Genes
This article digs into how the genetic variants you inherited from mom and dad influence the bacteria that can reside within you and how dietary changes can make a difference.
Emulsifiers in Processed Foods: Leaky Gut Genes
Emulsifiers, found in many processed and packaged foods, can trigger an inflammatory response in the intestines based on whether you carry specific genetic variants.
References:
Cellini, Barbara, et al. “The Chaperone Role of the Pyridoxal 5’-Phosphate and Its Implications for Rare Diseases Involving B6-Dependent Enzymes.” Clinical Biochemistry, vol. 47, no. 3, Feb. 2014, pp. 158–65. PubMed, https://doi.org/10.1016/j.clinbiochem.2013.11.021.
Chen, Lan X., and H. Ralph Schumacher. “192 – Other Crystal-Related Arthropathies.” Rheumatology (Sixth Edition), edited by Marc C. Hochberg et al., Mosby, 2015, pp. 1604–08. ScienceDirect, https://doi.org/10.1016/B978-0-323-09138-1.00192-3.
Coe, Fredric L., et al. “Kidney Stone Disease.” Journal of Clinical Investigation, vol. 115, no. 10, Oct. 2005, pp. 2598–608. PubMed Central, https://doi.org/10.1172/JCI26662.
Fargue, Sonia, et al. “Four of the Most Common Mutations in Primary Hyperoxaluria Type 1 Unmask the Cryptic Mitochondrial Targeting Sequence of Alanine:Glyoxylate Aminotransferase Encoded by the Polymorphic Minor Allele.” The Journal of Biological Chemistry, vol. 288, no. 4, Jan. 2013, pp. 2475–84. PubMed, https://doi.org/10.1074/jbc.M112.432617.
Garrelfs, Sander F., et al. “Patients with Primary Hyperoxaluria Type 2 Have Significant Morbidity and Require Careful Follow-Up.” Kidney International, vol. 96, no. 6, Dec. 2019, pp. 1389–99. ScienceDirect, https://doi.org/10.1016/j.kint.2019.08.018.
Grieff, Marvin, and David A. Bushinsky. “Chapter 42 – Nutritional Prevention and Treatment of Kidney Stones.” Nutritional Management of Renal Disease (Third Edition), edited by Joel D. Kopple et al., Academic Press, 2013, pp. 699–709. ScienceDirect, https://doi.org/10.1016/B978-0-12-391934-2.00042-4.
Gudbjartsson, Daniel F., et al. “Association of Variants at UMOD with Chronic Kidney Disease and Kidney Stones-Role of Age and Comorbid Diseases.” PLoS Genetics, vol. 6, no. 7, July 2010, p. e1001039. PubMed, https://doi.org/10.1371/journal.pgen.1001039.
“Home.” Journal of Osteopathic Medicine, https://jom.osteopathic.org/. Accessed 26 Aug. 2022.
How To Eat A Low Oxalate Diet | Kidney Stone Evaluation And Treatment Program. https://kidneystones.uchicago.edu/how-to-eat-a-low-oxalate-diet/. Accessed 26 Aug. 2022.
Hoyer-Kuhn, Heike, et al. “Vitamin B6 in Primary Hyperoxaluria I: First Prospective Trial after 40 Years of Practice.” Clinical Journal of the American Society of Nephrology: CJASN, vol. 9, no. 3, Mar. 2014, pp. 468–77. PubMed, https://doi.org/10.2215/CJN.06820613.
Kaufman, David W., et al. “Oxalobacter Formigenes May Reduce the Risk of Calcium Oxalate Kidney Stones.” Journal of the American Society of Nephrology : JASN, vol. 19, no. 6, June 2008, pp. 1197–203. PubMed Central, https://doi.org/10.1681/ASN.2007101058.
Köttgen, Anna, et al. “Uromodulin Levels Associate with a Common UMOD Variant and Risk for Incident CKD.” Journal of the American Society of Nephrology : JASN, vol. 21, no. 2, Feb. 2010, pp. 337–44. PubMed Central, https://doi.org/10.1681/ASN.2009070725.
Kumar, Vinod, et al. “Uromodulin Rs4293393 T>C Variation Is Associated with Kidney Disease in Patients with Type 2 Diabetes.” The Indian Journal of Medical Research, vol. 146, no. Suppl 2, Nov. 2017, pp. S15–21. PubMed Central, https://doi.org/10.4103/ijmr.IJMR_919_16.
Liebman, Michael, and Ismail A. Al-Wahsh. “Probiotics and Other Key Determinants of Dietary Oxalate Absorption1.” Advances in Nutrition, vol. 2, no. 3, Apr. 2011, pp. 254–60. PubMed Central, https://doi.org/10.3945/an.111.000414.
Lorenz, Elizabeth C., et al. “Update on Oxalate Crystal Disease.” Current Rheumatology Reports, vol. 15, no. 7, July 2013, p. 340. PubMed, https://doi.org/10.1007/s11926-013-0340-4.
Mitchell, Tanecia, et al. “Dietary Oxalate and Kidney Stone Formation.” American Journal of Physiology – Renal Physiology, vol. 316, no. 3, Mar. 2019, pp. F409–13. PubMed Central, https://doi.org/10.1152/ajprenal.00373.2018.
NM_012203.1(GRHPR):C.404+3_404+6delAAGT AND Primary Hyperoxaluria, Type II – ClinVar – NCBI. https://www.ncbi.nlm.nih.gov/clinvar/RCV000186457.2/. Accessed 26 Aug. 2022.
Piedra, María, et al. “Rs219780 SNP of Claudin 14 Gene Is Not Related to Clinical Expression in Primary Hyperparathyroidism.” Clinical Laboratory, vol. 61, no. 9, 2015, pp. 1197–203. PubMed, https://doi.org/10.7754/clin.lab.2015.150201.
“Primary Hyperoxaluria.” Wikipedia, 3 Aug. 2022. Wikipedia, https://en.wikipedia.org/w/index.php?title=Primary_hyperoxaluria&oldid=1102212898.
Santana, A., et al. “Primary Hyperoxaluria Type 1 in the Canary Islands: A Conformational Disease Due to I244T Mutation in the P11L-Containing Alanine:Glyoxylate Aminotransferase.” Proceedings of the National Academy of Sciences of the United States of America, vol. 100, no. 12, June 2003, pp. 7277–82. PubMed Central, https://doi.org/10.1073/pnas.1131968100.
Takayama, T., et al. “Ethnic Differences in GRHPR Mutations in Patients with Primary Hyperoxaluria Type 2: Ethnic Differences in GRHPR Mutations.” Clinical Genetics, vol. 86, no. 4, Oct. 2014, pp. 342–48. DOI.org (Crossref), https://doi.org/10.1111/cge.12292.
Thorleifsson, Gudmar, et al. “Sequence Variants in the CLDN14 Gene Associate with Kidney Stones and Bone Mineral Density.” Nature Genetics, vol. 41, no. 8, Aug. 2009, pp. 926–30. PubMed, https://doi.org/10.1038/ng.404.
Vezzoli, Giuseppe, Annalisa Terranegra, et al. “Calcium Kidney Stones Are Associated with a Haplotype of the Calcium-Sensing Receptor Gene Regulatory Region.” Nephrology, Dialysis, Transplantation: Official Publication of the European Dialysis and Transplant Association – European Renal Association, vol. 25, no. 7, July 2010, pp. 2245–52. PubMed, https://doi.org/10.1093/ndt/gfp760.
Vezzoli, Giuseppe, Alfredo Scillitani, et al. “Polymorphisms at the Regulatory Regions of the CASR Gene Influence Stone Risk in Primary Hyperparathyroidism.” European Journal of Endocrinology, vol. 164, no. 3, Mar. 2011, pp. 421–27. PubMed, https://doi.org/10.1530/EJE-10-0915.
Zimmermann, Diana J., et al. “Importance of Magnesium in Absorption and Excretion of Oxalate.” Urologia Internationalis, vol. 74, no. 3, 2005, pp. 262–67. PubMed, https://doi.org/10.1159/000083560.
Originally published Oct. 2015. Updated 3/2019.