Key takeaways:
~ Color vision is due to opsin genes, which encode proteins that are excited at different wavelengths of light.
~ Genetic mutations in these opsin genes give rise to different types of color blindness.
~ Differences in color vision may be offset by increased visual acuity.
~ You can check your 23 and Me or AncestryDNA genetic data for some of the mutations that cause color blindness.
Seeing in color:
Do we all see colors in the same way? Blue skies, green grass… may not look the same to everyone.
It turns out that variants in genes related to color blindness may mean we see things a bit differently.
When you see different colors, what you are really perceiving is different wavelengths of light.
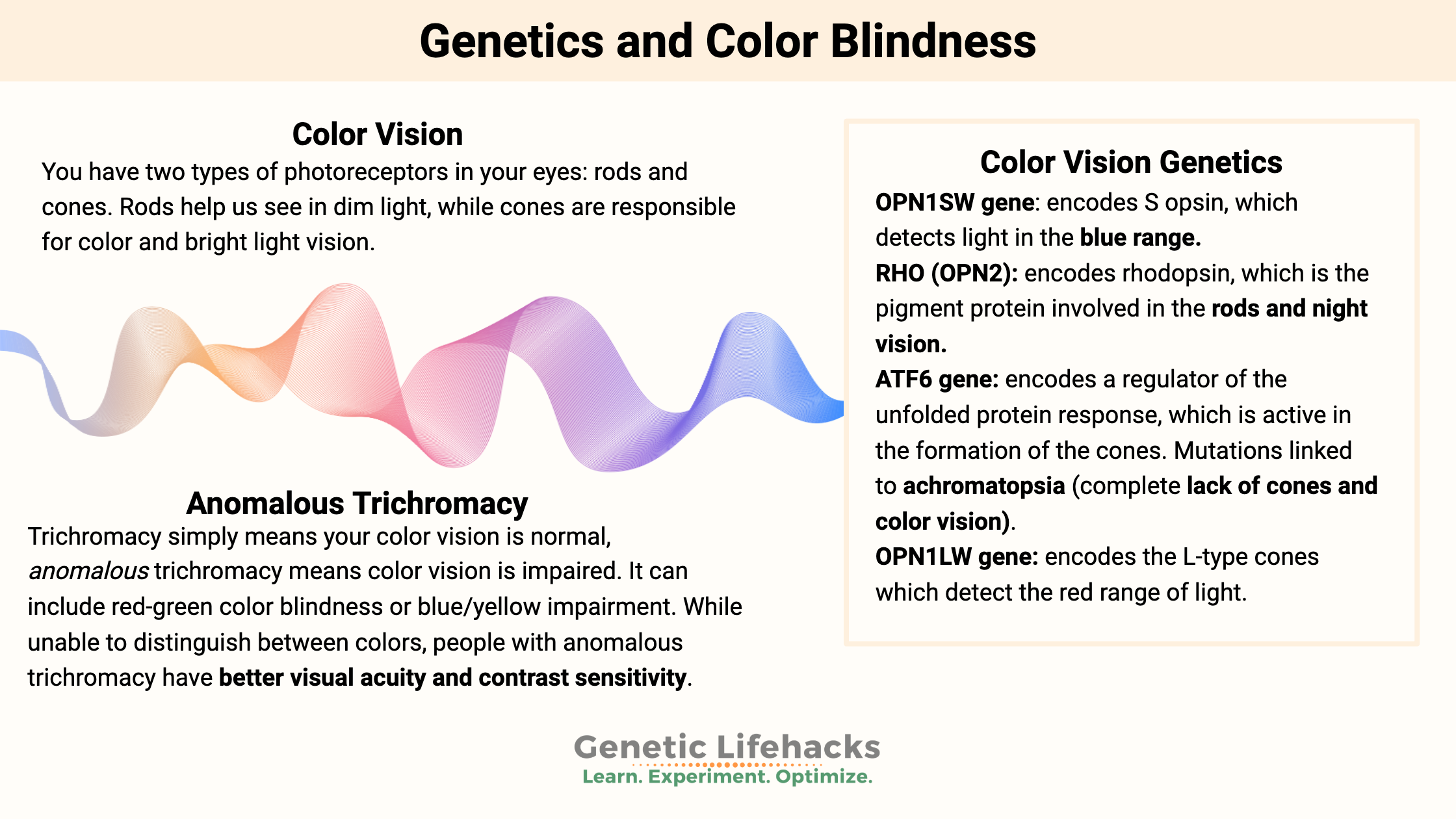
Rods and cones: giving you color and night vision
There are two types of photoreceptors in animals: rods and cones. Rods help us see in dim light, while cones are responsible for color and bright light vision.
Rods and cones use pigments for detecting light with different wavelengths. These pigments all end with the -opsin suffix, indicating they are a pigment:
- Rhodopsin is a pigment found in rods
- Several types of cone opsins are found in cones, detecting different colors or wavelengths of light.
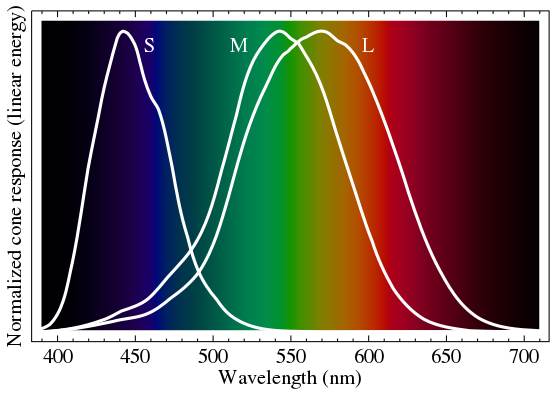
Short, medium, and long cone types help us distinguish colors from blue (short wavelengths) to green to red (long wavelengths).
Males only have one copy of the X chromosome, which contains the opsin genes. Due to this, color blindness is more likely to affect men than women when X-chromosome mutations occur.
Anomalous trichromacy: advantages and disadvantages
Trichromacy simply means your color vision is normal, with the three types of cone cells functioning.
Anomalous trichromacy means color vision is impaired. It can include red-green color blindness or blue/yellow impairment.
The impairment in detecting colors with any of the three types of cones can be more of a spectrum rather than a complete lack of seeing a color. A mild change in vision in a certain spectrum can lead to problems discriminating between colors.[article]
Deuteranomaly is a type of red-green color blindness where the green spectrum is not as sensitive, whereas, in protanomaly, the red spectrum is not as sensitive.
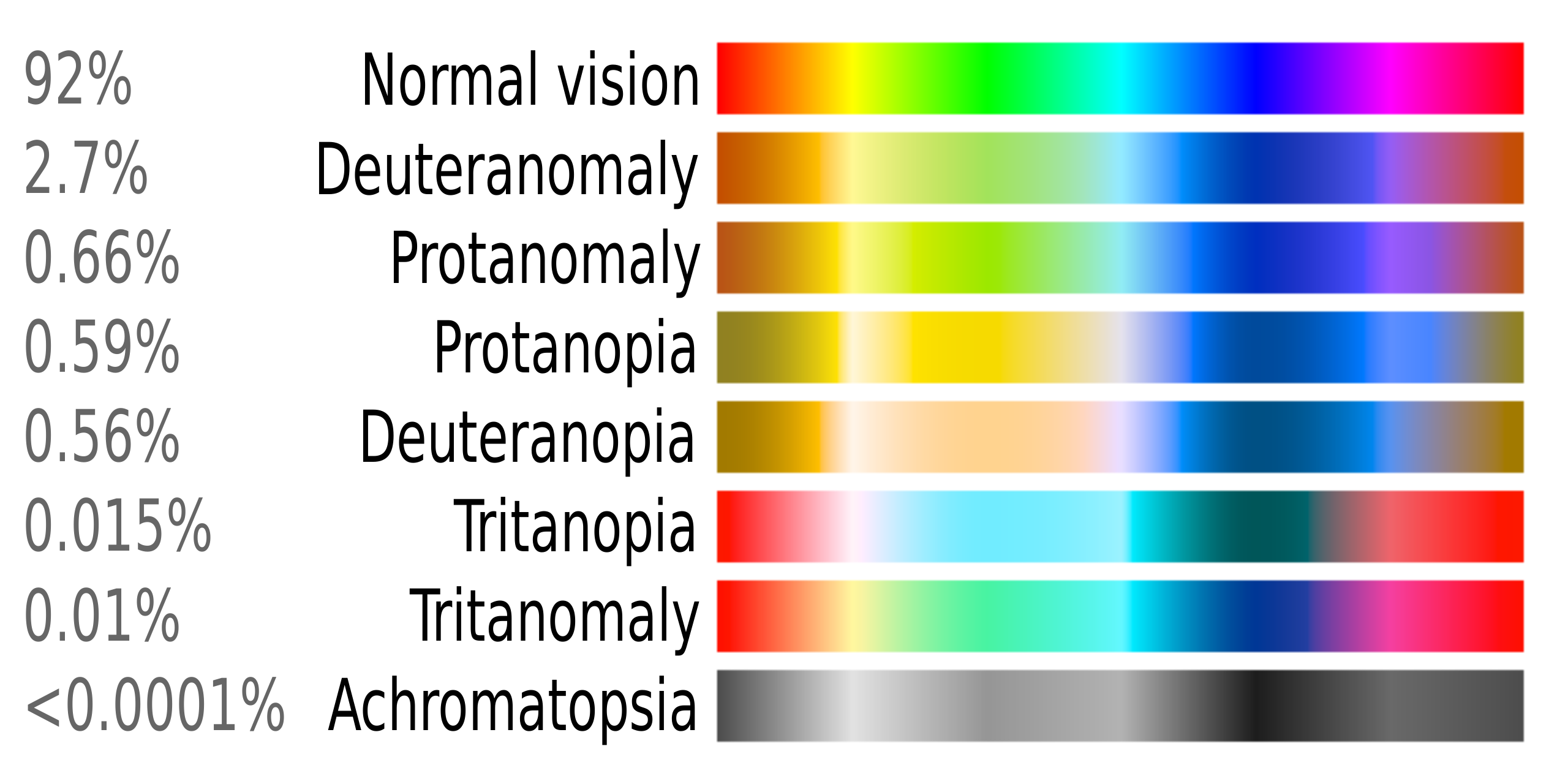
About 8% of Caucasian males have anomalous trichromacy, while in females, the problem affects around 1 in 200. (Think this applies to you? There’s a link in the Lifehacks section to how you can test this.)
It is a disadvantage in many ways to be unable to discriminate between certain colors. For example, people with red-green color blindness can’t pick out a ripe strawberry, and doctors who can’t see red have a hard time viewing rashes. There are more serious disadvantages when driving or operating machinery.
However, researchers have figured out there are also advantages to having color vision deficiencies.
For example, soldiers with color vision deficiencies can pick out someone in camouflage better than normal vision soldiers. Overall, people with anomalous trichromacy have better visual acuity and contrast sensitivity. It may explain why the mutations are so common in the human population — it was an immense advantage for part of the tribe to see camouflaged animals when hunting.[ref]
Recent fMRI studies show the brain compensates for decreased color sensitivity by amplifying the receptor signals.[ref]
Why are men more likely to be red-green color blind?
The red and green detecting genes are on the X chromosome, so men only have one copy of the chromosome, while women have two copies of the chromosome. Thus, mutations are going to show up dominantly in men.
Interestingly, most men with normal color vision have one L opsin gene and one -or more- copies of the M opsin gene. A complete lack of the L gene or the M gene can give rise to color blindness. Additionally, a combination of the L and M genes during the formation of the X chromosome can also occur, affecting color vision. About 10% of the population has more than three copies of the M opsin gene on the X chromosome.[ref]
Genotype Report for Color Blindness:
Lifehacks:
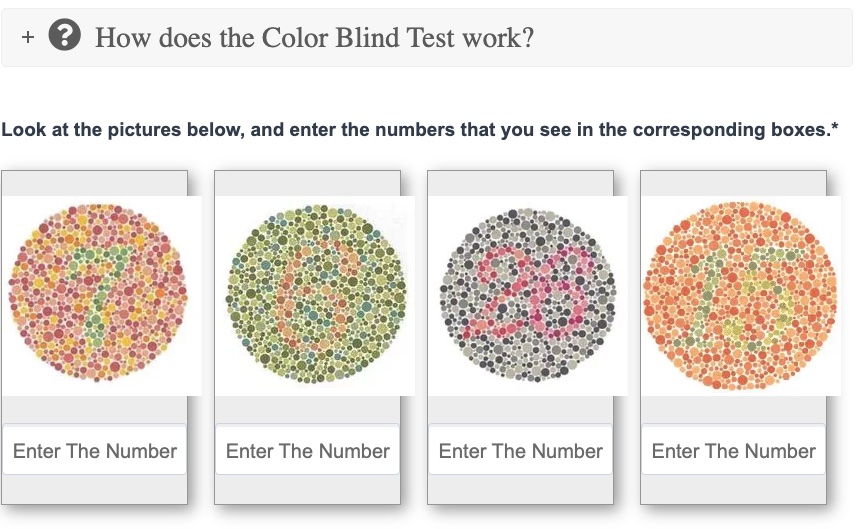
Take a color blindness test:
There are many color blindness tests online to see how well you discriminate between colors. Here’s one that’s free: https://colormax.org/color-blind-test/
Colored lenses:
Related Articles and Topics:
Trimethylaminuria: FMO3 mutations that cause malodorous body odor
References:
Baraas, Rigmor C., et al. “Substitution of Isoleucine for Threonine at Position 190 of S-Opsin Causes S-Cone-Function Abnormalities.” Vision Research, vol. 73, Nov. 2012, pp. 1–9. PubMed, https://doi.org/10.1016/j.visres.2012.09.007.
Carroll, Joseph, et al. “The Effect of Cone Opsin Mutations on Retinal Structure and the Integrity of the Photoreceptor Mosaic.” Investigative Ophthalmology & Visual Science, vol. 53, no. 13, Dec. 2012, pp. 8006–15. PubMed, https://doi.org/10.1167/iovs.12-11087.
“Color Blind Test | Test Your Color Vision | Ishihara Test for Color Blindness.” Color Vision Correction, https://colormax.org/color-blind-test/. Accessed 17 Oct. 2022.
Davidoff, Candice, et al. “Genetic Testing as a New Standard for Clinical Diagnosis of Color Vision Deficiencies.” Translational Vision Science & Technology, vol. 5, no. 5, Sept. 2016, p. 2. PubMed Central, https://doi.org/10.1167/tvst.5.5.2.
Doron, Ravid, et al. “Spatial Visual Function in Anomalous Trichromats: Is Less More?” PLoS ONE, vol. 14, no. 1, Jan. 2019, p. e0209662. PubMed Central, https://doi.org/10.1371/journal.pone.0209662.
Kohl, Susanne, et al. “Mutations in the Unfolded Protein Response Regulator ATF6 Cause the Cone Dysfunction Disorder Achromatopsia.” Nature Genetics, vol. 47, no. 7, July 2015, pp. 757–65. PubMed Central, https://doi.org/10.1038/ng.3319.
Neitz, Maureen, et al. “Tritan Color Vision Deficiency May Be Associated with an OPN1SW Splicing Defect and Haploinsufficiency.” Journal of the Optical Society of America. A, Optics, Image Science, and Vision, vol. 37, no. 4, Apr. 2020, pp. A26–34. PubMed, https://doi.org/10.1364/JOSAA.381919.
Perdices, Lorena, et al. “Systemic Epigallocatechin Gallate Protects against Retinal Degeneration and Hepatic Oxidative Stress in the P23H-1 Rat.” Neural Regeneration Research, vol. 17, no. 3, Mar. 2022, pp. 625–31. PubMed, https://doi.org/10.4103/1673-5374.320990.
Rodriguez-Carmona, Marisa, et al. “The Effects of Supplementation with Lutein and/or Zeaxanthin on Human Macular Pigment Density and Colour Vision.” Ophthalmic & Physiological Optics: The Journal of the British College of Ophthalmic Opticians (Optometrists), vol. 26, no. 2, Mar. 2006, pp. 137–47. PubMed, https://doi.org/10.1111/j.1475-1313.2006.00386.x.
Rs104893768 RefSNP Report – DbSNP – NCBI. https://www.ncbi.nlm.nih.gov/snp/rs104893768#clinical_significance. Accessed 17 Oct. 2022.
Tregillus, Katherine E. M., et al. “Color Compensation in Anomalous Trichromats Assessed with FMRI.” Current Biology: CB, vol. 31, no. 5, Mar. 2021, pp. 936-942.e4. PubMed, https://doi.org/10.1016/j.cub.2020.11.039.
Ueyama, Hisao, et al. “Novel Missense Mutations in Red/Green Opsin Genes in Congenital Color-Vision Deficiencies.” Biochemical and Biophysical Research Communications, vol. 294, no. 2, June 2002, pp. 205–09. PubMed, https://doi.org/10.1016/S0006-291X(02)00458-8.