Key takeaways:
~ Drinking alcohol is often a problem for people with histamine intolerance.
~ Two different genetic pathways are at play here: histamine degradation and/or alcohol metabolism pathways.
~ Understanding these pathways may help people with histamine sensitivity avoid alcohol-induced reactions.
Members will see their genotype report below, plus additional solutions in the Lifehacks section. Consider joining today.
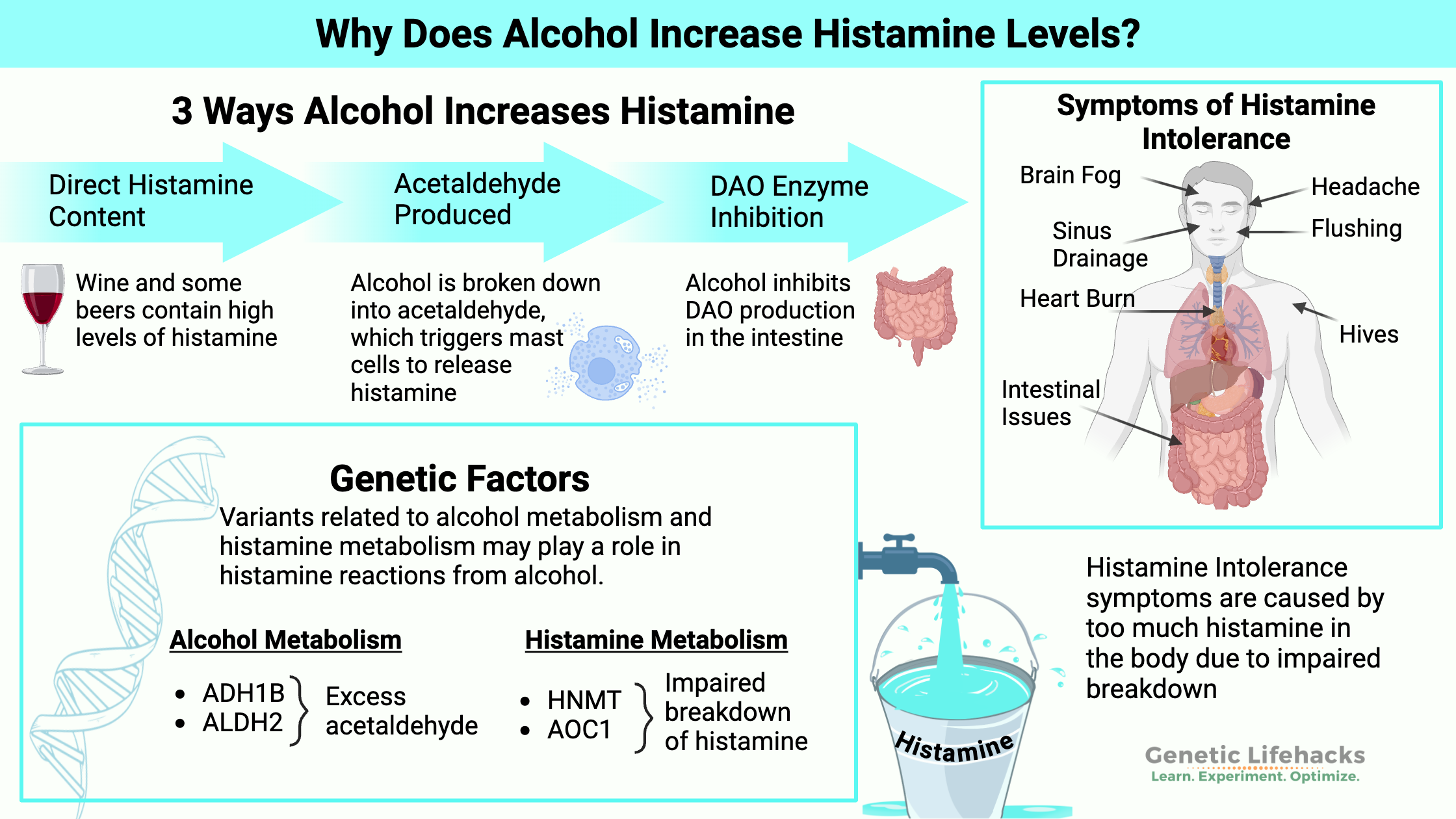
Why does alcohol increase histamine levels?
Drinking alcohol can trigger histamine reactions via several pathways.
- Alcoholic beverages often contain a lot of histamine. For people who don’t break down histamine well, this can cause a reaction.
- The breakdown of alcohol into acetaldehyde can trigger histamine release under certain conditions.
- Alcohol also inhibits the enzyme that breaks down histamine
First, I’ll explain a little of the background science on these pathways. Then, I’ll show you how you can use your genetic data to see which pathway is likely causing you problems.
What is histamine intolerance?
Histamine intolerance is a condition caused by the body’s inability to break down the histamine contained in foods and drinks. Normally, histamine from foods is broken down in the intestines.
Symptoms of histamine intolerance include headaches, itching, hives, sinus drainage, digestive issues, rapid heartbeat, anxiety, flushing, and (rarely) anaphylaxis.
How is histamine metabolized?
There are two ways your body clears histamine: the DAO enzyme or the HMNT enzyme.[ref]
- Diamine oxidase (DAO) enzyme:
Histamine from foods or bacteria in your gut is broken down or metabolized using the DAO (diamine oxidase) enzyme. The DAO enzyme is produced in the villi lining the small intestines and is released to metabolize histamine.[ref] - Histamine methyltransferase (HMNT) enzyme:
The HMNT enzyme works throughout the body, including in the brain, to deactivate and break down histamine created by your cells.[ref]
Too much histamine can result in allergy-like symptoms, including the histamine intolerance symptoms listed above.
Genetics comes into play here. Not producing enough DAO in the intestines can cause excess histamine throughout the body. Similarly, not enough HNMT enzyme can cause excess histamine in certain tissues.
Which foods are high in histamine? Fermented and aged foods are high in histamine — e.g., wine, aged cheeses, sausages, and processed meats. Other foods, such as strawberries, spinach, and chocolate, are also naturally higher in histamine. There’s a complete list in the Lifehacks section.
Histamine and biogenic amines in fermentation
Alcoholic beverages are made by fermentation, and histamine levels are generally high in fermented foods. For example, a 2009 study found all beers tested contained histamine.[ref]
Biogenic amines include histamine as well as tyramine, putrescine, cadaverine, and β-phenylethylamine. These biogenic amines can be produced by bacteria – specifically lactic acid producing bacteria used in fermentation.
Side note: Researchers think that lactic acid bacteria produce biogenic amines as a defense mechanism in order to withstand acidic environments.
Wine is fermented and is often very high in biogenic amines, including histamine. Levels of biogenic amines depend on the bacteria strains used in the fermentation process.[ref]
A recent Danish study found that 8% of the population had hypersensitive reactions to wine, including sinus drainage, sneezing, lower respiratory problems, or skin reactions (itching, hives).[ref]
However… Not all alcoholic beverages are high in histamine, so it isn’t as simple as all alcohol causes reactions due to containing histamine.
Alcohol exacerbates a lack of DAO
It is important to understand how alcohol can exacerbate symptoms if you suffer from histamine intolerance.
There are two ways alcohol can increase histamine:
- The DAO enzyme, which breaks down histamine, is inhibited by alcohol.[ref]
- Additionally, both alcohol and acetaldehyde can trigger mast cells and basophils to release histamine in the periphery. This increases histamine levels in different tissues, such as in the skin, causing itching and hives.[ref]
Metabolism of alcohol: creating acetaldehyde
I’ve mentioned acetaldehyde a couple of times without really explaining it…
Alcohol goes through a multistep process to be broken down and eliminated by the body.[ref]
- Alcohol is first metabolized by alcohol dehydrogenase (ADH genes) into acetaldehyde, which is toxic.
- Acetaldehyde is metabolized by acetaldehyde dehydrogenase (ALDH genes) into acetic acid, which is non-toxic.
In addition to alcohol-containing beverages, the alcohol dehydrogenase enzyme also breaks down alcohols produced by bacterial fermentation in the intestines. Retinol (vitamin A) and bile acids are also metabolized by alcohol dehydrogenase.[ref]

When alcohol is turned into acetaldehyde more quickly than the body can handle, there can be a buildup of acetaldehyde (toxic). Genetics plays a significant role in how quickly you turn alcohol into acetaldehyde.
Acetaldehyde needs to be quickly converted to acetic acid. Genetic variants that slow down this process cause a buildup of acetaldehyde.
As I mentioned above, excess acetaldehyde can trigger histamine release from mast cells and basophils.[ref]
Alcohol hypersensitivity
Doctors define alcohol hypersensitivity as an “exacerbation of respiratory symptoms in response to alcohol”.
A recent study of chronic rhinosinusitis patients with alcohol hypersensitivity found they also had higher platelet counts. In these patients, red wine extract activated the platelet-adherent basophils. This is important because basophils release histamine when activated.[ref]
Alcohol can also trigger asthma reactions in some people. Asthma is an airway hyperactivation involving histamine.
In people with reduced ALDH2 levels (and thus more acetaldehyde), alcohol is more likely to induce asthma.[ref]
One study explains: “Impaired ALDH2 activity leads to high peak levels of acetaldehyde and to histamine release.” Studies show that inhaling acetaldehyde induces histamine-mediated airway constriction, such as is seen in asthma.[ref]
Hangovers and histamine
A recent animal study sheds light on the age-old question of what causes hangover symptoms.
Researchers found some hangover symptoms (headache, behavioral inhibition, dizziness, movement imbalance) are caused by high histamine levels. Treating the high histamine levels resulted in decreasing levels of specific migraine-related neurotransmitters.[ref]
Histamine and Alcohol Genotype Report
Lifehacks:
Let’s talk about some ways of avoiding histamine reactions from alcohol!
Low histamine diet and alcohol:
If you are not on a low histamine diet already and are noticing histamine reactions from alcohol, you may want to consider avoiding high-histamine foods when drinking alcohol.
Think of it this way: If you have already eaten a bunch of foods high in histamine, your body is still working on degrading the histamine from food. Going low-histamine for a day or two before you’re going to be drinking alcohol may help your body have enough of the necessary enzymes on hand to break down the histamine in the alcoholic beverage.
A low-histamine diet restricts foods with high levels of histamine and foods that cause the body to release histamine. To experiment with a low-histamine diet, eliminate all of the higher-histamine foods for a period of time to see how your body responds.
In general, fermented and aged foods are higher in histamine. A quick overview of high histamine foods includes processed meats, cheeses (except farmer cheese), and seafood that isn’t completely fresh. Foods naturally high in histamine include spinach, chocolate, tomatoes, strawberries, wine, sake, and more.
Pizza and beer peeps: Pepperoni pizza with marinara sauce is usually very high in histamine. This may answer the question as to why beer and pizza give you problems.
Wine and cheese crowd: Similarly, if you are a wine and cheese lover, aged cheeses are extremely high in histamine. Avoiding the cheese, meat, and olive tray and instead opting for carrots and celery off the veggie tray may help your histamine reactions.
What to avoid: If you are considering a low histamine diet, I find this printable histamine food list to be the most comprehensive: Complete list of foods high in histamine (pdf).
Quit drinking…
Simply put, avoiding alcohol altogether is the easiest way to avoid histamine reactions from alcohol. Keep in mind that non-alcoholic beer and alcohol-free wines may still contain histamine.
Low histamine wines?
New wines are on the market that claim to be low in histamine. Natural wines that contain only grapes are likely to be lower in histamine, in part because they don’t include histamine releasing preservatives such as sulfur dioxide. Generally, white wines are usually lower than red. [ref]
Histamine-targeting supplements:
Related Articles and Topics:
Histamine Intolerance: Genetic Report, Supplements, and Real Solutions
References:
Agúndez, José A. G., et al. “The Diamine Oxidase Gene Is Associated with Hypersensitivity Response to Non-Steroidal Anti-Inflammatory Drugs.” PLoS ONE, vol. 7, no. 11, Nov. 2012, p. e47571. PubMed Central, https://doi.org/10.1371/journal.pone.0047571.
Che, Denis Nchang, et al. “Fisetin Inhibits IL-31 Production in Stimulated Human Mast Cells: Possibilities of Fisetin Being Exploited to Treat Histamine-Independent Pruritus.” Life Sciences, vol. 201, May 2018, pp. 121–29. PubMed, https://doi.org/10.1016/j.lfs.2018.03.056.
Crabb, David W., et al. “Overview of the Role of Alcohol Dehydrogenase and Aldehyde Dehydrogenase and Their Variants in the Genesis of Alcohol-Related Pathology.” The Proceedings of the Nutrition Society, vol. 63, no. 1, Feb. 2004, pp. 49–63. PubMed, https://doi.org/10.1079/pns2003327.
Edenberg, Howard J. “The Genetics of Alcohol Metabolism: Role of Alcohol Dehydrogenase and Aldehyde Dehydrogenase Variants.” Alcohol Research & Health, vol. 30, no. 1, 2007, pp. 5–13. PubMed Central, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3860432/.
Ehlers, Cindy L. “Variations in ADH and ALDH in Southwest California Indians.” Alcohol Research & Health, vol. 30, no. 1, 2007, pp. 14–17. PubMed Central, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3860438/.
Eschenbacher, William, et al. “Activation of Platelet-Adherent Basophils in Chronic Rhinosinusitis with Alcohol Hypersensitivity.” Annals of Allergy, Asthma & Immunology: Official Publication of the American College of Allergy, Asthma, & Immunology, vol. 128, no. 4, Apr. 2022, pp. 443–50. PubMed, https://doi.org/10.1016/j.anai.2022.01.013.
García-Martín, Elena, Carmen Martínez, et al. “Diamine Oxidase Rs10156191 and Rs2052129 Variants Are Associated with the Risk for Migraine.” Headache, vol. 55, no. 2, Feb. 2015, pp. 276–86. PubMed, https://doi.org/10.1111/head.12493.
García-Martín, Elena, Pedro Ayuso, et al. “Histamine Pharmacogenomics.” Pharmacogenomics, vol. 10, no. 5, May 2009, pp. 867–83. PubMed, https://doi.org/10.2217/pgs.09.26.
Hao, Liuyi, et al. “Mitochondria-Targeted Ubiquinone (MitoQ) Enhances Acetaldehyde Clearance by Reversing Alcohol-Induced Posttranslational Modification of Aldehyde Dehydrogenase 2: A Molecular Mechanism of Protection against Alcoholic Liver Disease.” Redox Biology, vol. 14, Apr. 2018, pp. 626–36. PubMed, https://doi.org/10.1016/j.redox.2017.11.005.
Hon, Yuen Yi, et al. “Endogenous Histamine and Cortisol Levels in Subjects with Different Histamine N-Methyltransferase C314T Genotypes.” Molecular Diagnosis & Therapy, vol. 10, no. 2, 2006, pp. 109–14. PubMed Central, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4178529/.
—. “Endogenous Histamine and Cortisol Levels in Subjects with Different Histamine N-Methyltransferase C314T Genotypes : A Pilot Study.” Molecular Diagnosis & Therapy, vol. 10, no. 2, 2006, pp. 109–14. PubMed, https://doi.org/10.1007/BF03256450.
Jones, Bridgette L., et al. “Genetic Variation in the Histamine Production, Response, and Degradation Pathway Is Associated with Histamine Pharmacodynamic Response in Children with Asthma.” Frontiers in Pharmacology, vol. 7, Jan. 2017, p. 524. PubMed Central, https://doi.org/10.3389/fphar.2016.00524.
Kempuraj, D., et al. “Luteolin Inhibits Myelin Basic Protein-Induced Human Mast Cell Activation and Mast Cell-Dependent Stimulation of Jurkat T Cells.” British Journal of Pharmacology, vol. 155, no. 7, Dec. 2008, pp. 1076–84. PubMed Central, https://doi.org/10.1038/bjp.2008.356.
Langhi, Cédric, et al. “Regulation of Human Class I Alcohol Dehydrogenases by Bile Acids.” Journal of Lipid Research, vol. 54, no. 9, Sept. 2013, pp. 2475–84. PubMed, https://doi.org/10.1194/jlr.M039404.
Maintz, L., et al. “Association of Single Nucleotide Polymorphisms in the Diamine Oxidase Gene with Diamine Oxidase Serum Activities.” Allergy, vol. 66, no. 7, July 2011, pp. 893–902. PubMed, https://doi.org/10.1111/j.1398-9995.2011.02548.x.
Matsuse, H., et al. “Screening for Acetaldehyde Dehydrogenase 2 Genotype in Alcohol-Induced Asthma by Using the Ethanol Patch Test.” The Journal of Allergy and Clinical Immunology, vol. 108, no. 5, Nov. 2001, pp. 715–19. PubMed, https://doi.org/10.1067/mai.2001.118791.
Meza-Velázquez, R., et al. “Association of Diamine Oxidase and Histamine N-Methyltransferase Polymorphisms with Presence of Migraine in a Group of Mexican Mothers of Children with Allergies.” Neurología (English Edition), vol. 32, no. 8, Oct. 2017, pp. 500–07. ScienceDirect, https://doi.org/10.1016/j.nrleng.2016.02.012.
Park, Hyo-Hyun, Soyoung Lee, Jae-Min Oh, et al. “Anti-Inflammatory Activity of Fisetin in Human Mast Cells (HMC-1).” Pharmacological Research, vol. 55, no. 1, Jan. 2007, pp. 31–37. PubMed, https://doi.org/10.1016/j.phrs.2006.10.002.
Park, Hyo-Hyun, Soyoung Lee, Hee-Young Son, et al. “Flavonoids Inhibit Histamine Release and Expression of Proinflammatory Cytokines in Mast Cells.” Archives of Pharmacal Research, vol. 31, no. 10, Oct. 2008, pp. 1303–11. PubMed, https://doi.org/10.1007/s12272-001-2110-5.
“PDB101: Molecule of the Month: Alcohol Dehydrogenase.” RCSB: PDB-101, http://pdb101.rcsb.org/motm/13. Accessed 20 Dec. 2022.
Raje, Nikita, et al. “Genetic Variation within the Histamine Pathway among Patients with Asthma.” The Journal of Asthma : Official Journal of the Association for the Care of Asthma, vol. 52, no. 4, May 2015, pp. 353–62. PubMed Central, https://doi.org/10.3109/02770903.2014.973501.
Regecová, Ivana, et al. “Detection of Microbiota during the Fermentation Process of Wine in Relation to the Biogenic Amine Content.” Foods, vol. 11, no. 19, Oct. 2022. www.ncbi.nlm.nih.gov, https://doi.org/10.3390/foods11193061.
Shimoda, Terufumi, et al. “The Pathogenesis of Alcohol-Induced Airflow Limitation in Acetaldehyde Dehydrogenase 2-Deficient Mice.” International Archives of Allergy and Immunology, vol. 171, no. 3–4, 2016, pp. 276–84. www.karger.com, https://doi.org/10.1159/000452709.
Shulpekova, Yulia O., et al. “Food Intolerance: The Role of Histamine.” Nutrients, vol. 13, no. 9, Sept. 2021, p. 3207. PubMed Central, https://doi.org/10.3390/nu13093207.
Tang, Tao, et al. “Determination of Biogenic Amines in Beer with Pre-Column Derivatization by High Performance Liquid Chromatography.” Journal of Chromatography. B, Analytical Technologies in the Biomedical and Life Sciences, vol. 877, no. 5–6, Feb. 2009, pp. 507–12. PubMed, https://doi.org/10.1016/j.jchromb.2008.12.064.
Thompson, Jon S. “Significance of the Intestinal Gradient of Diamine Oxidase Activity.” Digestive Diseases, vol. 8, no. 3, 1990, pp. 163–68. www.karger.com, https://doi.org/10.1159/000171249.
Weng, Zuyi, et al. “Quercetin Is More Effective than Cromolyn in Blocking Human Mast Cell Cytokine Release and Inhibits Contact Dermatitis and Photosensitivity in Humans.” PloS One, vol. 7, no. 3, 2012, p. e33805. PubMed, https://doi.org/10.1371/journal.pone.0033805.
Wüthrich, B. “Allergic and Intolerance Reactions to Wine.” Allergologie Select, vol. 2, no. 1, Sept. 2018, pp. 80–88. PubMed Central, https://doi.org/10.5414/ALX01420E.
Yokoyama, Akira, et al. “Combinations of Alcohol-Induced Flushing with Genetic Polymorphisms of Alcohol and Aldehyde Dehydrogenases and the Risk of Alcohol Dependence in Japanese Men and Women.” PLoS ONE, vol. 16, no. 7, July 2021, p. e0255276. PubMed Central, https://doi.org/10.1371/journal.pone.0255276.
Yoshikawa, Takeo, et al. “Histamine N-Methyltransferase in the Brain.” International Journal of Molecular Sciences, vol. 20, no. 3, Feb. 2019, p. 737. PubMed Central, https://doi.org/10.3390/ijms20030737.
Zhao, Wenmei, et al. “The Effects of Biogenic Amines in Chinese Huangjiu on the Behavior of Mice and Hangover Headache‐related Indices.” Food Science & Nutrition, vol. 10, no. 12, Aug. 2022, pp. 4226–37. PubMed Central, https://doi.org/10.1002/fsn3.3016.
Zimatkin, S. M., and O. V. Anichtchik. “Alcohol-Histamine Interactions.” Alcohol and Alcoholism (Oxford, Oxfordshire), vol. 34, no. 2, 1999, pp. 141–47. PubMed, https://doi.org/10.1093/alcalc/34.2.141.