Key takeaways:
~ Celiac disease is an autoimmune disorder in which gluten causes damage to the small intestines.
~ Celiac is only possible in people with specific HLA genotypes.
~ You can use your genetic raw data (23andMe, AncestryDNA, etc) to find out if you have the HLA type that makes you susceptible to celiac.
Members will see their genotype report below and the solutions in the Lifehacks section. Consider joining today.
Members will see their genotype report below, plus additional solutions in the Lifehacks section. Consider joining today.
What is celiac disease?
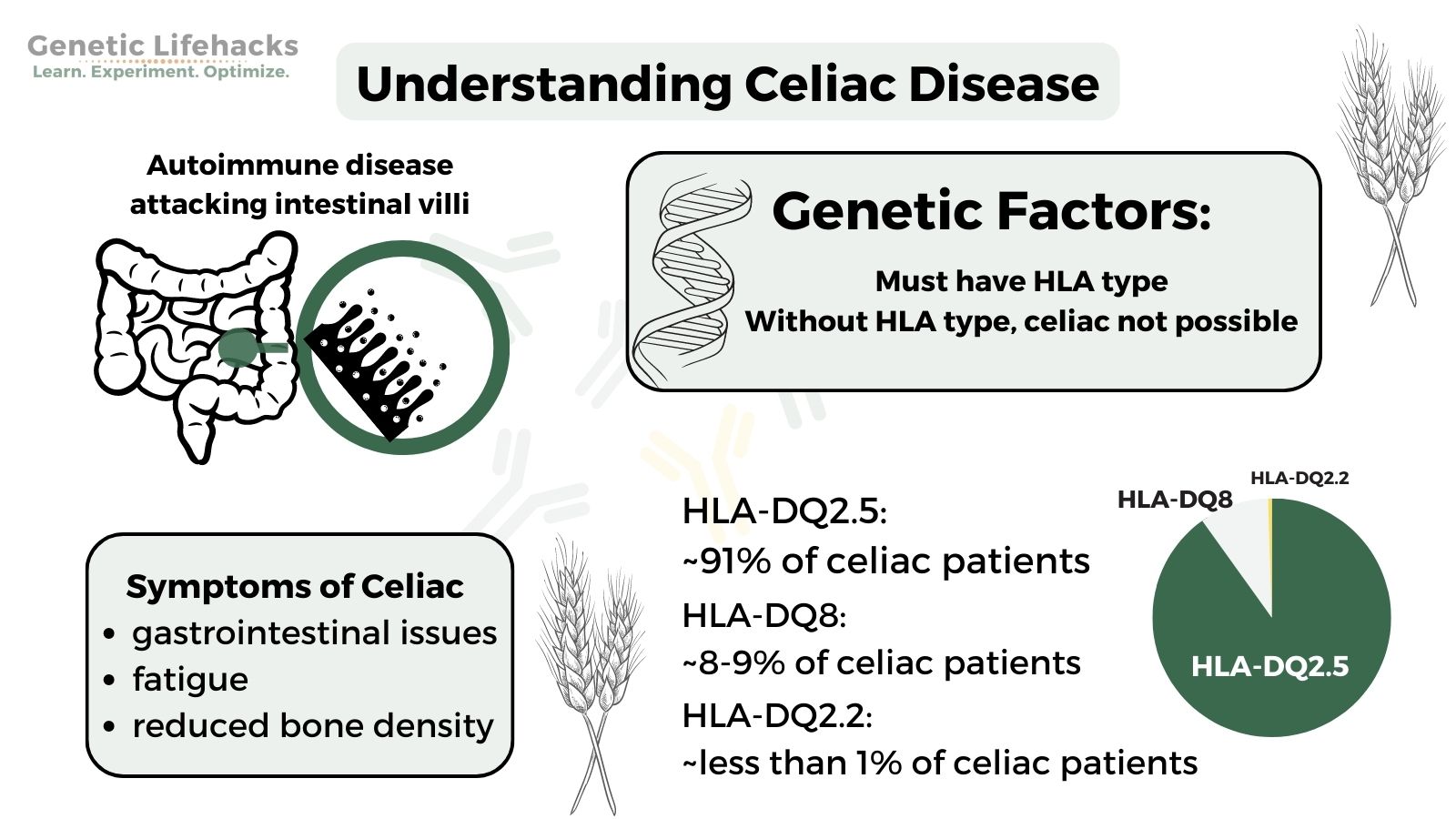
Celiac disease (spelled coeliac in Britain) is an autoimmune disorder in which gluten, a protein found in wheat and other grains, leads to your body attacking its own cells.
Inside the small intestines, there are little projections called villi. In celiac disease, the body attacks those cells, causing the villi to shorten and not absorb nutrients very well.
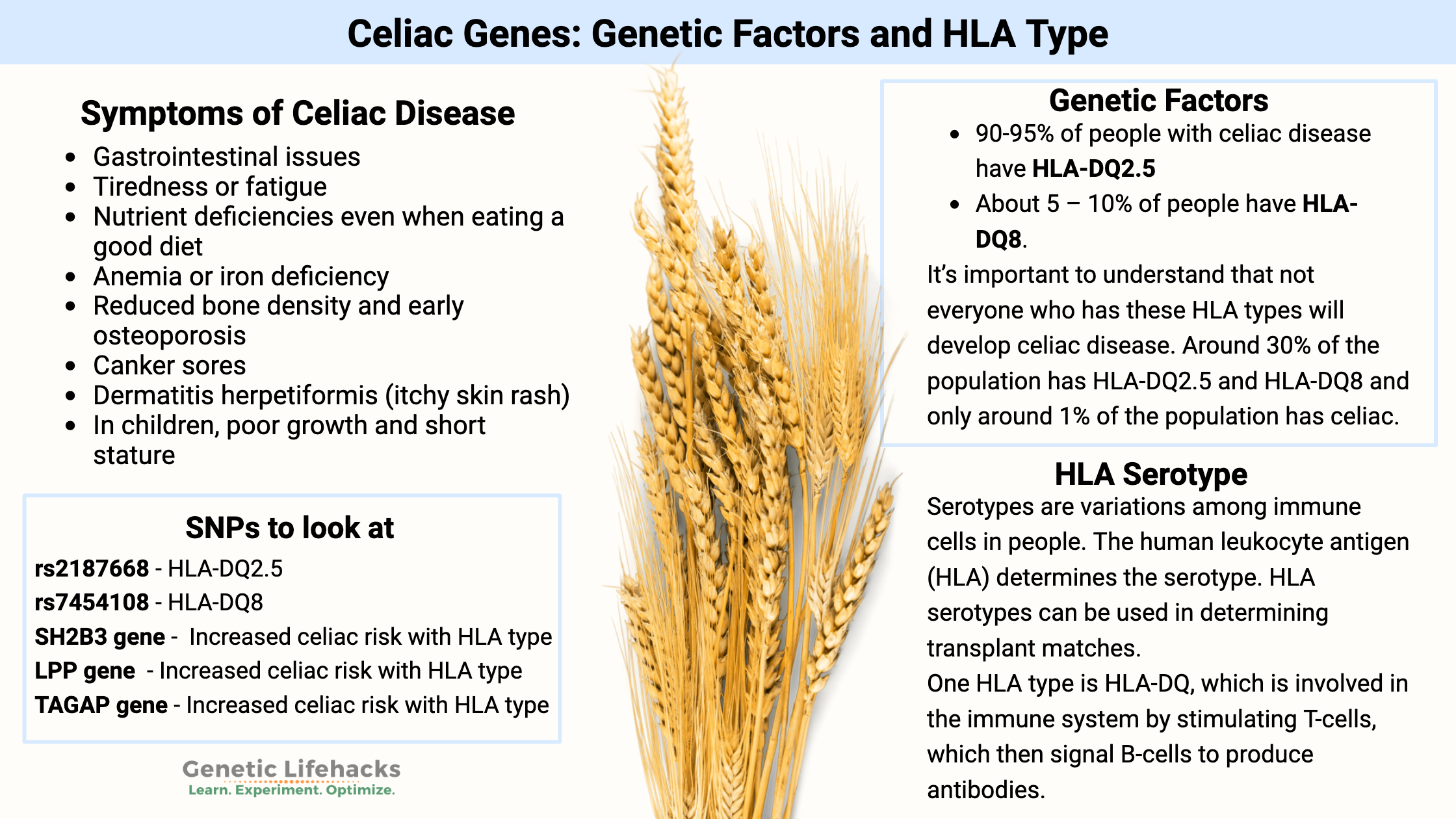
Symptoms of celiac disease:
Symptoms of celiac disease can vary from person to person, but some common ones include: [ref]
- Gastrointestinal issues
- Tiredness or fatigue
- Nutrient deficiencies even when eating a good diet
- Anemia or iron deficiency
- Reduced bone density and early osteoporosis
- Canker sores
- Dermatitis herpetiformis (itchy skin rash)
- In children, poor growth and short stature
It’s also worth noting that not everyone with celiac disease will have all of these symptoms.
Is celiac disease hereditary?
Genetic diseases that are hereditary are caused by inheriting a genetic variant or mutation from your parents.
While there isn’t just one specific genetic variant that causes everyone who carries it to get celiac disease, it is hereditary and based on genetic variants.
In order for a person to be susceptible to celiac disease, certain genetic variants must be present. If these specific variants, called HLA alleles, are not present, it is highly unlikely that a person has celiac disease.
Almost everyone with Celiac disease has either the HLA-DQ2 or HLA-DQ8 alleles.
- 90-95% of people with celiac disease have HLA-DQ2.5
- About 5 – 10% of people have HLA-DQ8.
- Some studies also list HLA-DQ2.2 as a possibility for celiac, with about 1% of celiac patients carrying HLA-DQ2.2.
It’s important to understand that not everyone who has these HLA types will develop celiac disease. In fact, almost 25% of the population has HLA-DQ2.5, and that percentage grows to ~30% when adding in HLA-DQ8 alleles.[ref]
With only ~1% of the population having celiac disease, you can see that just having the HLA type doesn’t mean that you will get celiac disease.
Thus, looking at your genetic variants can help you rule out celiac disease, but it cannot tell you if you have it.
What is an HLA serotype?
Serotypes, discovered in 1933 by Rebecca Lancefield, are variations within species (bacteria and viruses) or variations among immune cells in people. The typing is based on their cell surface antigens. In humans, the human leukocyte antigen (HLA) determines the serotype. HLA serotypes can be used in determining transplant matches.
One HLA type is HLA-DQ, which is a protein found on antigen-presenting cells. DQ is involved in the immune system by stimulating T-cells, which then signal B-cells to produce antibodies.
HLA-DQ recognizes foreign antigens from pathogens, but it also recognizes common self-antigens. This is where the problem begins when the HLA-DQ loses its tolerance to self-proteins, triggering autoimmune diseases such as celiac, lupus, and type 1 diabetes.
Diagnosing Celiac Disease:
There are blood tests that can show whether you carry antibodies against gluten. The test is not 100% accurate, and some people may have false-negative results (e.g., a blood test shows that you don’t have celiac when you do).
Genetics can help you rule out celiac disease. If you don’t carry the HLA type (below) and also have a negative antibody test, then celiac is highly unlikely.
The ‘gold standard’ is an intestinal biopsy, where a gastroenterologist takes a small snip out of your intestines to see if there is damage to the villi.
Celiac Genotype Report
Members: Log in to see your data below.
Not a member? Join here.
Why is this section is now only for members? Here’s why…
Lifehacks:
The rest of this article is for Genetic Lifehacks members only. Consider joining today to see the rest of this article.
Related Articles and Topics:
Diabetes: Genetic Risk Report
We often talk about diabetes as though it is one disease, but diabetes can have several different causes or pathways that are impacting glucose regulation. Tailoring your diabetes prevention (or reversal) efforts to fit your genetic susceptibility may be more effective.
Thyroid Hormone Levels and Your Genes
Your genes play a big role in how well your thyroid works and how your body produces and converts the different forms of thyroid hormone. Genetic variants can impact your risk for hypothyroidism, Hashimoto’s, and Graves’ disease.
Mast cells: MCAS, genetics, and solutions
Mast Cell Activation Syndrome, or MCAS, is a recently recognized disease involving mast cells that are misbehaving in various ways. Symptoms of MCAS can include abdominal pain, nausea, itching, flushing, hives, headaches, heart palpitations, anxiety, brain fog, and anaphylaxis.
Problems with IBS? Personalized solutions based on your genes
There are multiple causes of IBS, and genetics can play a role in IBS symptoms. Pinpointing your cause can help you to figure out your solution.
Graphical Abstract:
References:
Canova, Cristina, et al. “Celiac Disease and Risk of Autoimmune Disorders: A Population-Based Matched Birth Cohort Study.” The Journal of Pediatrics, vol. 174, July 2016, pp. 146-152.e1. PubMed, https://doi.org/10.1016/j.jpeds.2016.02.058.
Huang, Shi-Qi, et al. “Association of LPP and TAGAP Polymorphisms with Celiac Disease Risk: A Meta-Analysis.” International Journal of Environmental Research and Public Health, vol. 14, no. 2, Feb. 2017, p. 171. PubMed Central, https://doi.org/10.3390/ijerph14020171.
Hunt, Karen A., et al. “Newly Identified Genetic Risk Variants for Celiac Disease Related to the Immune Response.” Nature Genetics, vol. 40, no. 4, Apr. 2008, pp. 395–402. PubMed, https://doi.org/10.1038/ng.102.
Izzo, Valentina, et al. “Improving the Estimation of Celiac Disease Sibling Risk by Non-HLA Genes.” PLoS ONE, vol. 6, no. 11, Nov. 2011, p. e26920. PubMed Central, https://doi.org/10.1371/journal.pone.0026920.
—. “Improving the Estimation of Celiac Disease Sibling Risk by Non-HLA Genes.” PloS One, vol. 6, no. 11, 2011, p. e26920. PubMed, https://doi.org/10.1371/journal.pone.0026920.
—. “Improving the Estimation of Celiac Disease Sibling Risk by Non-HLA Genes.” PLoS ONE, vol. 6, no. 11, Nov. 2011, p. e26920. PubMed Central, https://doi.org/10.1371/journal.pone.0026920.
—. “Improving the Estimation of Celiac Disease Sibling Risk by Non-HLA Genes.” PLoS ONE, vol. 6, no. 11, Nov. 2011, p. e26920. PubMed Central, https://doi.org/10.1371/journal.pone.0026920.
Leccioli, Valentina, et al. “A New Proposal for the Pathogenic Mechanism of Non-Coeliac/Non-Allergic Gluten/Wheat Sensitivity: Piecing Together the Puzzle of Recent Scientific Evidence.” Nutrients, vol. 9, no. 11, Nov. 2017, p. 1203. PubMed Central, https://doi.org/10.3390/nu9111203.
Megiorni, Francesca, and Antonio Pizzuti. “HLA-DQA1 and HLA-DQB1 in Celiac Disease Predisposition: Practical Implications of the HLA Molecular Typing.” Journal of Biomedical Science, vol. 19, no. 1, Oct. 2012, p. 88. BioMed Central, https://doi.org/10.1186/1423-0127-19-88.
Monsuur, Alienke J., et al. “Effective Detection of Human Leukocyte Antigen Risk Alleles in Celiac Disease Using Tag Single Nucleotide Polymorphisms.” PLoS ONE, vol. 3, no. 5, May 2008, p. e2270. PubMed Central, https://doi.org/10.1371/journal.pone.0002270.
Parmar, A. S., et al. “Association Study of FUT2 (Rs601338) with Celiac Disease and Inflammatory Bowel Disease in the Finnish Population.” Tissue Antigens, vol. 80, no. 6, Dec. 2012, pp. 488–93. PubMed, https://doi.org/10.1111/tan.12016.
“Symptoms of Celiac Disease.” Celiac Disease Foundation, https://celiac.org/about-celiac-disease/symptoms-of-celiac-disease/. Accessed 16 Dec. 2021.
Ting, Yi Tian, et al. “A Molecular Basis for the T Cell Response in HLA-DQ2.2 Mediated Celiac Disease.” Proceedings of the National Academy of Sciences of the United States of America, vol. 117, no. 6, Feb. 2020, pp. 3063–73. PubMed Central, https://doi.org/10.1073/pnas.1914308117.
Trynka, Gosia, et al. “Dense Genotyping Identifies and Localizes Multiple Common and Rare Variant Association Signals in Celiac Disease.” Nature Genetics, vol. 43, no. 12, Nov. 2011, pp. 1193–201. PubMed Central, https://doi.org/10.1038/ng.998.
Tye-Din, J. A., et al. “Appropriate Clinical Use of Human Leukocyte Antigen Typing for Coeliac Disease: An Australasian Perspective.” Internal Medicine Journal, vol. 45, no. 4, Apr. 2015, pp. 441–50. PubMed Central, https://doi.org/10.1111/imj.12716.
Yang, Guang, et al. “Systematic Review and Meta-Analysis of the Association between IL18RAP Rs917997 and CCR3 Rs6441961 Polymorphisms with Celiac Disease Risks.” Expert Review of Gastroenterology & Hepatology, vol. 9, no. 10, 2015, pp. 1327–38. PubMed, https://doi.org/10.1586/17474124.2015.1075880.