When you first start learning about how your genes affect your health, it can be overwhelming. What is truly important? Are there any SNPs in your 23andMe data that are actually interesting?
Let me help you cut through the confusion with my list of what is important to check first. The following are ten genes with significant variants that can seriously impact health. Members will see their genotype report below, plus additional solutions in the Lifehacks section. Consider joining today.
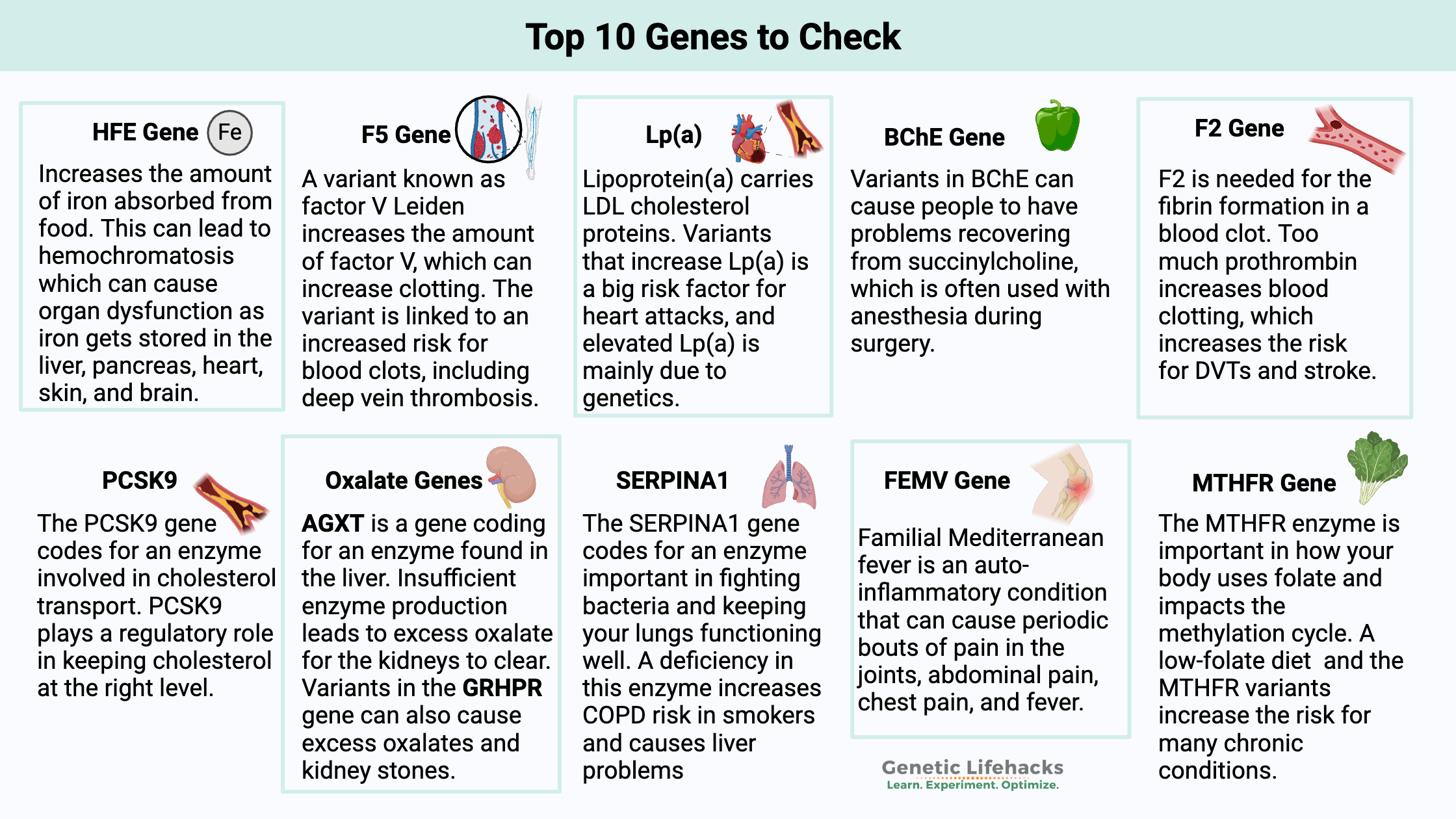
Top 10 List: Genes and SNPs To Check in Your Raw Data
If you have one of the risk variants listed below, be sure to click through to read the full article for in-depth information and the listed ‘Lifehacks’ for that gene. Understanding your genetic susceptibility can guide you towards preventing future diseases.
Not a member? Join here. Membership lets you see your data right in the genotype report below.
Top 10 Genes Genotype Report:
#1) HFE Gene (hemochromatosis risk)
A couple of mutations in the HFE gene increase the amount of iron absorbed from food. It causes increased iron storage in various tissues in the body. Eventually, this can lead to hemochromatosis or iron overload. Symptoms of too much iron can include joint pain (often moving from joint to joint) and fatigue. Hemochromatosis can involve organ dysfunction as iron gets stored in the liver, pancreas, heart, skin, and brain. Read the full article on iron overload
Check your genetic data for rs1800562 (23andMe v4, v5; AncestryDNA):
- A/A: most common cause of hereditary hemochromatosis, highest ferritin levels (two copies of C282Y)
- A/G: increased ferritin levels can cause hemochromatosis. Population frequency of about 5% in people with Caucasian/European heritage.
- G/G: typical
Members: Your genotype for rs1800562 is —.
Check your genetic data for rs1799945 (23andMe v4, v5; AncestryDNA):
- G/G: can cause mild hemochromatosis, increased ferritin levels (two copies of H63D)
- C/G: somewhat higher ferritin levels, usually not a bit problem unless combined with C282Y. Population frequency of about 14% in people with Caucasian/European heritage.
- C/C: typical
Members: Your genotype for rs1799945 is —.
#2) F5 Gene (factor V Leiden blood clot risk)
Factor V is a clotting factor essential for forming blood clots. A gene variant known as factor V Leiden increases the amount of factor V, which can increase clotting. The variant is linked to an increased risk for blood clots, including deep vein thrombosis. Read the full article on factor V Leiden.
Check your genetic data for rs6025 (23andMe v4, v5; AncestryDNA):
- C/C: typical
- C/T: one copy of factor V Leiden, significantly increased risk of blood clots. Population frequency of about 5% in Caucasians, slightly lower in other population groups.
- T/T: two copies of factor V Leiden, significantly increased risk of blood clots
Members: Your genotype for rs6025 is —.
#3) Lp(a) (heart attack risk)
Lipoprotein(a) carries LDL cholesterol proteins. Elevated Lp(a) is a big risk factor for heart attacks, and elevated Lp(a) is mainly due to genetics. Check out the full article on lipoprotein(a).
Check your genetic data for rs3798220 (23andMe v4, v5,AncestryDNA):
- C/C: risk of elevated Lp(a), significantly increased risk for heart disease – 3.7x risk of aortic stenosis[ref][ref]
- C/T: risk of elevated Lp(A), increased risk for heart disease, and increased aortic stenosis risk. Global population frequency of about 15%.
- T/T: typical
Members: Your genotype for rs3798220 is —.
Check your genetic data for rs10455872 (23andMe v4, v5; AncestryDNA):
- G/G: likely elevated Lp(a), significantly increased risk for heart disease – 2x risk of aortic stenosis[ref][ref]
- A/G: likely elevated Lp(A), increased risk for heart disease
- A/A: typical
Members: Your genotype for rs10455872 is —.
#4) BChE Gene (A-variant and anesthesia)
The BChE enzyme is vital for breaking down certain toxins affecting acetylcholine, an important neurotransmitter. Mutations in BChE can cause people to have problems recovering from succinylcholine, which is often used with anesthesia during surgery. Read the full article on BChE.
Check your genetic data for rs1799807 (23andMe v4, v5; AncestryDNA):
- T/T: typical
- C/T: one copy of A-variant, may have delayed recovery from succinylcholine[ref][ref][ref][ref]. Population frequency of about 1% in Caucasians/Europeans. Rare in other population groups.
- C/C: two copies of A-variant (very rare), severe BChE deficiency
Members: Your genotype for rs1799807 is —.
#5) F2 Gene (Prothrombin, blood clot risk)
Prothrombin (or factor 2) is part of the blood-clotting cascade of events. It is needed for the fibrin formation in a blood clot. Too much prothrombin increases blood clotting, which increases the risk for DVTs. Read the full prothrombin article.
Check your genetic data for rs1799963, G20210A (23andMe i3002432; AncestryDNA):
- A/A: a large increase in the risk of blood clots, increased risk of stroke with PFO[ref]
- A/G: increased risk of blood clots, increased risk of stroke with PFO
- G/G: typical
Members: Your genotype for i3002432 / rs1799963 is .
#6) PCSK9 (high cholesterol)
The PCSK9 gene codes for an enzyme involved in cholesterol transport. The enzyme binds to LDL particles, which transport fat molecules, including cholesterol, throughout the body. PCSK9 plays a regulatory role in keeping cholesterol at the right level. Check out the full article on PCSK9.
PCSK9 variants associated with increased LDL-cholesterol:
Check your genetic data for rs505151 (23andMe v4, v5; AncestryDNA):
- G/G: increased LDL, increased risk of coronary artery disease[ref][ref][ref]
- A/G: somewhat increased LDL, increased risk of coronary artery disease. About 20% of the population globally carries this risk allele.
- A/A: typical
Members: Your genotype for rs505151 is —.
Check your genetic data for rs28942112 (23andMe i5000370, v4; AncestryDNA):
- C/T: greatly increased LDL, considered pathogenic for familial hypercholesterolemia[ref]. It is a rare mutation.
- T/T: typical
Members: Your genotype for rs28942112 (i5000370) is .
#7) Oxalate Genes (kidney stones due to oxalates)
AGXT (alanine-glyoxylate aminotransferase) is a gene coding for an enzyme found in the liver. It helps convert a form of oxalate made by the body (glyoxylate) into glycine. Insufficient enzyme production leads to excess oxalate for the kidneys to clear. Mutations in the GRHPR gene can also cause excess oxalates and kidney stones. Read the full article on oxalates.
Check your genetic data for rs34116584 (23andMe v4 only):
- T/T: significantly increases the risk of hyperoxaluria[ref][ref]
- C/T: increased risk of hyperoxaluria (especially if coupled with another mutation)[ref]. Over 10% of the population carries this variant.
- C/C: typical
Members: Your genotype for rs34116584 is —.
Check your genetic data for rs180177309 (23andMe i5012629 v4, v5):
- II or AAGT/AAGT: typical
- DI or -/AAGT: carrier of a pathogenic allele for primary hyperoxaluria type 2
- DD or -/-: primary hyperoxaluria type 2[ref]
Members: Your genotype for rs180177309 is —.
Check your genetic data for rs80356708 (23andMe i5012628 v4, v5):
- II (or G/G): typical
- DI (or – / G): carrier of a pathogenic allele for primary hyperoxaluria type 2
- DD (or -/-): primary hyperoxaluria type 2[ref]
Members: Your genotype for rs80356708 is —.
#8) SERPINA1 Gene (Alpha-1 Antitrypsin Deficiency)
The SERPINA1 gene codes for an enzyme important in fighting bacteria and keeping your lungs functioning well. A deficiency in this enzyme increases COPD risk in smokers and causes liver problems, especially in people who drink alcohol. Read the full article on Alpha-1 Antitrypsin Deficiency.
Check your genetic data for rs28929474 (23andMe v4, v5; AncestryDNA *):
- T/T: two copies of Pi*Z mutation, alpha-1 antitrypsin levels often less than 30% of normal[ref]
- C/T: one copy of the Pi*Z mutation
- C/C: typical
Members: Your genotype for rs28929474 is —.
Check your genetic data for rs17580 (23andMe v4, v5; AncestryDNA *):
- A/A: two copies of Pi*S mutation
- A/T: one copy of the Pi*S mutation
- T/T: typical
Members: Your genotype for rs17580 is —.
*All rs id data is given in the plus orientation to match 23andMe and AncestryDNA genetic raw data format.
#9) FEMV Gene (familial Mediterranean fever)
Familial Mediterranean fever is an auto-inflammatory condition that can cause periodic bouts of pain in the joints, abdominal pain, chest pain, and fever. Not everyone who carries the mutations will end up with periodic episodes. Read the full article on familial Mediterranean fever.
Check your genetic data for rs61732874 (23andMe i4000409 v4, v5; AncestryDNA):
- A/A: FMF variant A744S (two copies)
- A/C: one copy of FMF variant
- C/C: typical
Members: Your genotype for rs61732874 is —.
Check your genetic data for rs3743930 (23andMe v4, v5; AncestryDNA):
- G/G: FMF variant E148Q (two copies)
- C/G: one copy of FMF variant
- C/C: typical
Members: Your genotype for rs3743930 is —.
Check your genetic data for rs104895083 (23andMe i4000403 v4, v5):
- C/C: FMF variant F479L (two copies)
- C/G: one copy of FMF variant
- G/G: typical
Members: Your genotype for rs104895083 is —.
Check your genetic data for rs104895094 (23andMe i4000407 v4, v5):
- C/C: FMF variant K695R (two copies)
- C/T: one copy of FMF variant
- T/T: typical
Members: Your genotype for i4000407 is —.
Check your genetic data for rs28940580 (23andMe v4, v5):
- G/G: FMF variant M680I (two copies)
- C/G: one copy of FMF variant
- C/C: typical
Members: Your genotype for rs28940580 is —.
Check your genetic data for rs28940578 (23andMe v4, v5; AncestryDNA):
- T/T: FMF variant M694I (two copies)
- C/T: one copy of FMF variant
- C/C: typical
Members: Your genotype for rs28940578 is —.
Check your genetic data for rs61752717 (23andMe i4000406 v4, v5):
- C/C: FMF variant M694V (two copies)
- C/T: one copy of FMF variant
- T/T: typical
Members: Your genotype for rs61752717 is —.
Check your genetic data for rs11466023 (23andMe v4, v5):
- A/A: FMF variant P369S (two copies)
- A/G: one copy of FMF variant
- G/G: typical
Members: Your genotype for rs11466023 is —.
Check your genetic data for rs104895097 (23andMe i4000410 v4, v5 ; AncestryDNA):
- T/T: FMF variant R761H (two copies)
- C/T: one copy of FMF variant
- C/C: typical
Members: Your genotype for rs104895097 is —.
Check your genetic data for rs28940579 (23andMe v4, v5; AncestryDNA):
- G/G: FMF variant V726A (two copies)
- A/G: one copy of FMF variant
- A/A: typical
Members: Your genotype for rs28940579 is —.
#10 MTHFR Gene (Methylation cycle):
The MTHFR enzyme is important in how your body uses folate and impacts the methylation cycle. A low-folate diet (not eating your veggies?) and the MTHFR variants increase the risk for many chronic conditions. Read the full article on MTHFR.
Check your genetic data for rs1801133 (23andMe v4, v5; AncestryDNA):
- G/G: typical
- A/G: one copy of MTHFR C677T allele (heterozygous), decreased by 40%
- A/A: two copies of MTHFR C677T (homozygous), decreased by 70 – 80%
Members: Your genotype for rs1801133 is —.
Check your genetic data for rs1801131 (23andMe v4, v5; AncestryDNA):
- T/T: typical
- G/T: one copy of MTHFR A1298C (heterozygous), slightly decreased enzyme function
- G/G: two copies of MTHFR A1298C (homozygous), decreased enzyme by about 20%
Members: Your genotype for rs1801131 is —.
Wait! What about Alzheimer’s risk??
Alzheimer’s risk is a touchy subject for some people, so I didn’t want to throw it in here. If you are interested, I would encourage you to read the full article on Alzheimer’s risk if you want to know this information.
References:
Chasman, Daniel I., et al. “Polymorphism in the Apolipoprotein(a) Gene, Plasma Lipoprotein(a), Cardiovascular Disease, and Low-Dose Aspirin Therapy.” Atherosclerosis, vol. 203, no. 2, Apr. 2009, pp. 371–76. PubMed, https://doi.org/10.1016/j.atherosclerosis.2008.07.019.
Chen, Suet N., et al. “A Common PCSK9 Haplotype, Encompassing the E670G Coding Single Nucleotide Polymorphism, Is a Novel Genetic Marker for Plasma Low-Density Lipoprotein Cholesterol Levels and Severity of Coronary Atherosclerosis.” Journal of the American College of Cardiology, vol. 45, no. 10, May 2005, pp. 1611–19. PubMed, https://doi.org/10.1016/j.jacc.2005.01.051.
Erhart, Gertraud, et al. “Genetic Factors Explain a Major Fraction of the 50% Lower Lipoprotein(a) Concentrations in Finns.” Arteriosclerosis, Thrombosis, and Vascular Biology, vol. 38, no. 5, May 2018, pp. 1230–41. PubMed Central, https://doi.org/10.1161/ATVBAHA.118.310865.
Fargue, Sonia, et al. “Four of the Most Common Mutations in Primary Hyperoxaluria Type 1 Unmask the Cryptic Mitochondrial Targeting Sequence of Alanine:Glyoxylate Aminotransferase Encoded by the Polymorphic Minor Allele.” The Journal of Biological Chemistry, vol. 288, no. 4, Jan. 2013, pp. 2475–84. PubMed, https://doi.org/10.1074/jbc.M112.432617.
Gomes, Henrique J. P., et al. “Investigation of Association between Susceptibility to Leprosy and SNPs inside and near the BCHE Gene of Butyrylcholinesterase.” Journal of Tropical Medicine, vol. 2012, 2012, p. 184819. PubMed, https://doi.org/10.1155/2012/184819.
Lando, Giuliana, et al. “Frequency of Butyrylcholinesterase Gene Mutations in Individuals with Abnormal Inhibition Numbers: An Italian-Population Study.” Pharmacogenetics, vol. 13, no. 5, May 2003, pp. 265–70. PubMed, https://doi.org/10.1097/00008571-200305000-00005.
Laschkolnig, Anja, et al. “Lipoprotein (a) Concentrations, Apolipoprotein (a) Phenotypes, and Peripheral Arterial Disease in Three Independent Cohorts.” Cardiovascular Research, vol. 103, no. 1, July 2014, pp. 28–36. PubMed, https://doi.org/10.1093/cvr/cvu107.
NM_000055.2(BCHE):C.293A>G (p.Asp98Gly) AND Deficiency of Butyrylcholine Esterase – ClinVar – NCBI. https://www.ncbi.nlm.nih.gov/clinvar/RCV000277104.1/. Accessed 19 May 2022.
NM_012203.1(GRHPR):C.404+3_404+6delAAGT AND Primary Hyperoxaluria, Type II – ClinVar – NCBI. https://www.ncbi.nlm.nih.gov/clinvar/RCV000186457.2/. Accessed 19 May 2022.
Pezzini, Alessandro, et al. “Do Common Prothrombotic Mutations Influence the Risk of Cerebral Ischaemia in Patients with Patent Foramen Ovale? Systematic Review and Meta-Analysis.” Thrombosis and Haemostasis, vol. 101, no. 5, May 2009, pp. 813–17.
Qiu, Chengfeng, et al. “What Is the Impact of PCSK9 Rs505151 and Rs11591147 Polymorphisms on Serum Lipids Level and Cardiovascular Risk: A Meta-Analysis.” Lipids in Health and Disease, vol. 16, no. 1, June 2017, p. 111. PubMed, https://doi.org/10.1186/s12944-017-0506-6.
Slimani, Afef, et al. “Effect of E670G Polymorphism in PCSK9 Gene on the Risk and Severity of Coronary Heart Disease and Ischemic Stroke in a Tunisian Cohort.” Journal of Molecular Neuroscience: MN, vol. 53, no. 2, June 2014, pp. 150–57. PubMed, https://doi.org/10.1007/s12031-014-0238-2.
Thun, Gian Andri, et al. “Causal and Synthetic Associations of Variants in the SERPINA Gene Cluster with Alpha1-Antitrypsin Serum Levels.” PLoS Genetics, vol. 9, no. 8, Aug. 2013, p. e1003585. PubMed Central, https://doi.org/10.1371/journal.pgen.1003585.
Zhu, Guang-dan, et al. “Genetic Testing for BCHE Variants Identifies Patients at Risk of Prolonged Neuromuscular Blockade in Response to Succinylcholine.” Pharmacogenomics and Personalized Medicine, vol. 13, Sept. 2020, pp. 405–14. PubMed Central, https://doi.org/10.2147/PGPM.S263741.